Diagnostic method for quickly detecting drug resistance gene CMY-2 of germ AmpC enzyme
A CMY-2 and drug-resistant gene technology, applied in the direction of recombinant DNA technology, DNA / RNA fragments, etc., can solve the problems of severe prevalence of plasmid AmpC enzyme, lack of detection methods, outbreaks of hospital infection, etc., and achieve high specificity and specificity Good performance and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 For Rapid Detection of Bacterial AmpC Enzyme Drug Resistance Gene CMY-2 Establishment of the LAMP kit
[0033] LAMP kit for rapid detection of bacterial extended-spectrum β-lactamase resistance gene PME-1, including the following components: (1) specific primer set; (2) DNA polymerase; (3) LAMP reaction solution; (4) Chromogenic reagent; (5) Positive control and negative control.
[0034] (1) Design of specific primers:
[0035] According to bacterial AmpC enzyme resistance gene CMY-2 (GenBank accession number is: (DQ173299)) Specifically designed 6 specific primers, the sequences are as follows:
[0036] Internal primer 1: 5'-CCTGGTAGATAACGGCAACGGTCGTTAATCGCACCATCAC-3' (SEQ ID No.1);
[0037] Internal primer 2: 5'- AGCCGATATCGCCAATAACCACACGTCTTACTAACCGATCCT-3' (SEQ ID No.2);
[0038] Outer primer 1: 5'- AGAACAACAGATTGCCGAT-3' (SEQ ID No.3);
[0039]Outer primer 2: 5'- GGCCAGTATTTCGTGACC-3' (SEQ ID No.4);
[0040] Loop primer 1: 5'-CGGAATAGCCTGCTCCTG-...
Embodiment 2
[0046] Example 2 Bacterial AmpC Enzyme Drug Resistance Gene CMY-2 rapid detection method
[0047] Utilize the kit of embodiment 1 to rapidly detect bacterial AmpC enzyme drug-resistant gene CMY-2 ,Specific steps are as follows:
[0048] (1) Extract template DNA: Extract bacterial genomic DNA by boiling water;
[0049] (2) Loop-mediated isothermal gene amplification reaction: 25 μL reaction system contains: inner primers 1 and 2 at a final concentration of 8 pmol / μL each, outer primers 1 and 2 at a final concentration of 1 pmol / μL each, loop primers 1 and 2 The final concentration is 4 pmol / μL, the reaction solution is 12.5 μL, the DNA polymerase is 8U, and the DNA to be tested is 50 ng. Make up to 25 μL with sterilized deionized water, then add 20 μL of sealing solution, and put the above reaction tube at 63 °C. Reaction 60min;
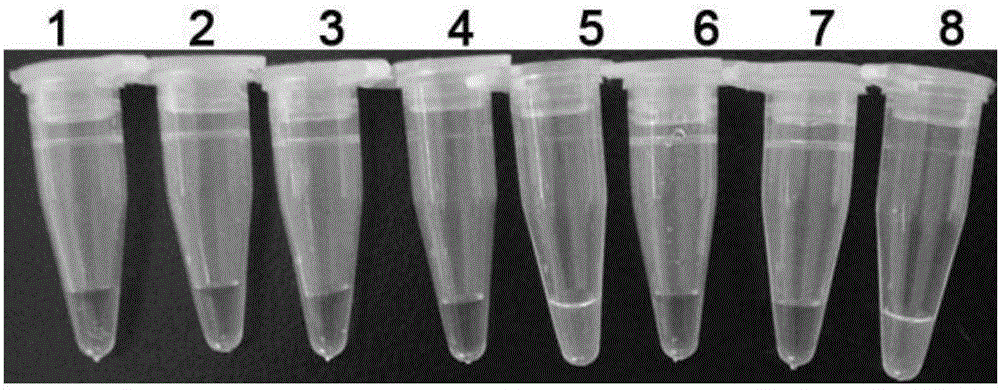
[0050] (3) Judgment of results: Add 1 μL of chromogen 1×SYBR Green I to the above reaction tube, and observe the color of the reaction solution...
Embodiment 3
[0051] Embodiment 3 specificity experiment
[0052] Utilize the method of embodiment 2 to identify not containing CMY-2 Gene clinical isolates were tested, and DEPC water was used as a negative control.
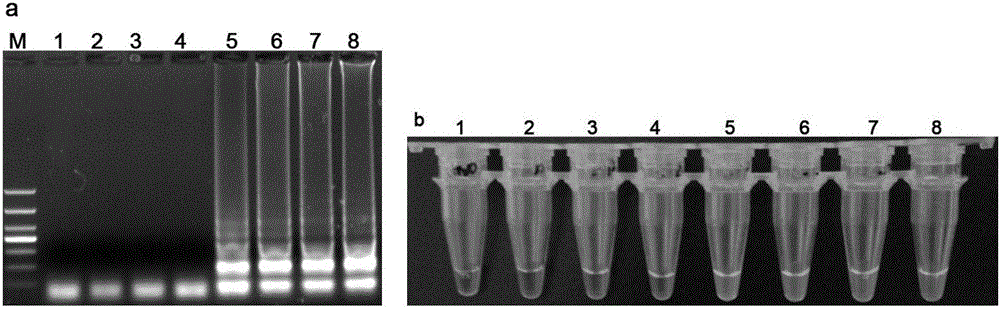
[0053] See the test results figure 2 . Bacterial AmpC enzyme resistance gene only CMY-2 The tube is green and the rest of the tubes are orange. The results show that the detection kit of the present invention has high specificity and can accurately identify bacterial AmpC enzyme drug resistance gene CMY-2 and other non-bacterial AmpC enzyme resistance genes CMY-2 differentiate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com