Single gene mRNA (messenger ribonucleic acid) methylation level detection method
A level detection and methylation technology, applied in the field of molecular biology, can solve the problems of time-consuming and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Total RNA extraction:
[0042] Take 50-100mg of two groups of pig liver samples and perform the following operations: after grinding in liquid nitrogen, add 1ml Trizol lysate and mix well, add 0.2ml chloroform and shake vigorously for 15 seconds, incubate at room temperature for 10min, then centrifuge at 4°C and 15000g for 15min , transfer the upper aqueous phase (about 500 μl) to a new tube, add 500 μl of isopropanol, mix upside down; centrifuge at 15,000 g for 10 min at 4°C, discard the supernatant; add 1ml of 75% (volume %) ethanol to wash, 4°C , Centrifuge at 15,000g for 10min, discard the supernatant, and when the RNA precipitate at the bottom of the tube becomes colorless and transparent, add 80μl of DEPC water and blow to mix.
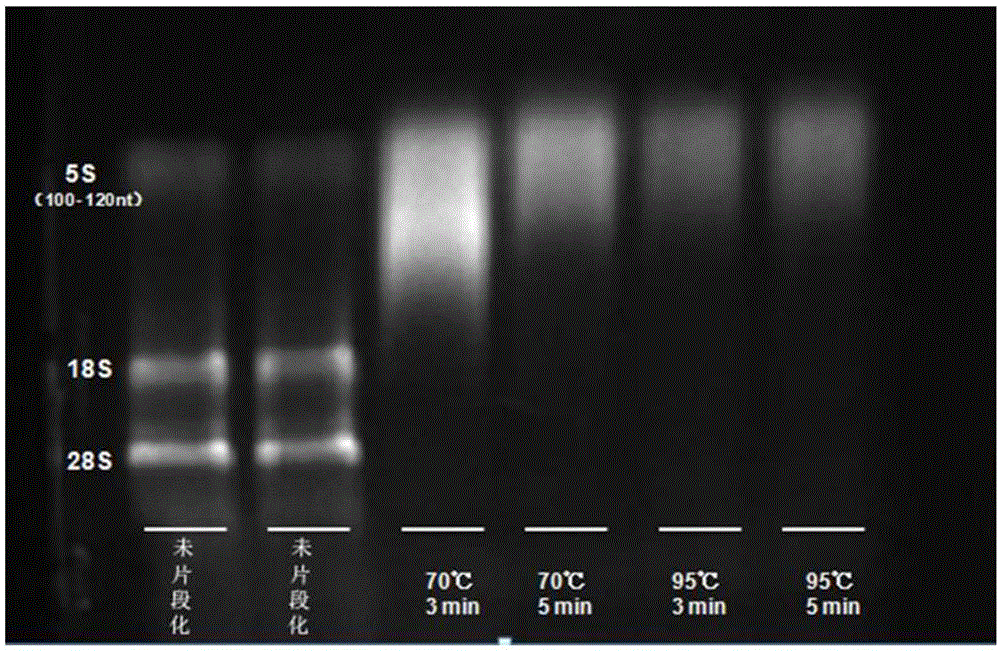
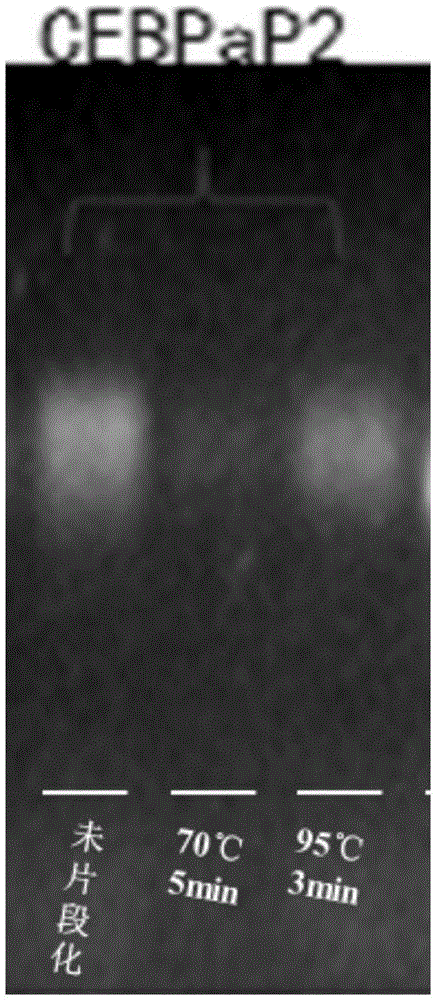
[0043] 2. RNA fragmentation:
[0044] 1) Mix the RNA obtained in step 1 with the fragmentation buffer according to the following reaction system,
[0045] RNA Fragmentation System
[0046]
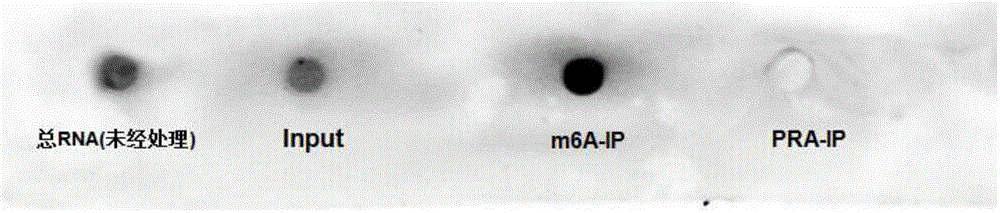
[0047] In order to obtain the optimal ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com