In-vitro gene stacking technology compatible with recombinase-mediated in-vivo gene stacking and application of in-vitro gene stacking technology
A technology of gene stacking and recombinase, which is applied in the direction of recombinant DNA technology, introduction of foreign genetic material using vectors, etc., can solve the problems of increasing examination costs and time, and achieve the effect of fast preparation method and easy application.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
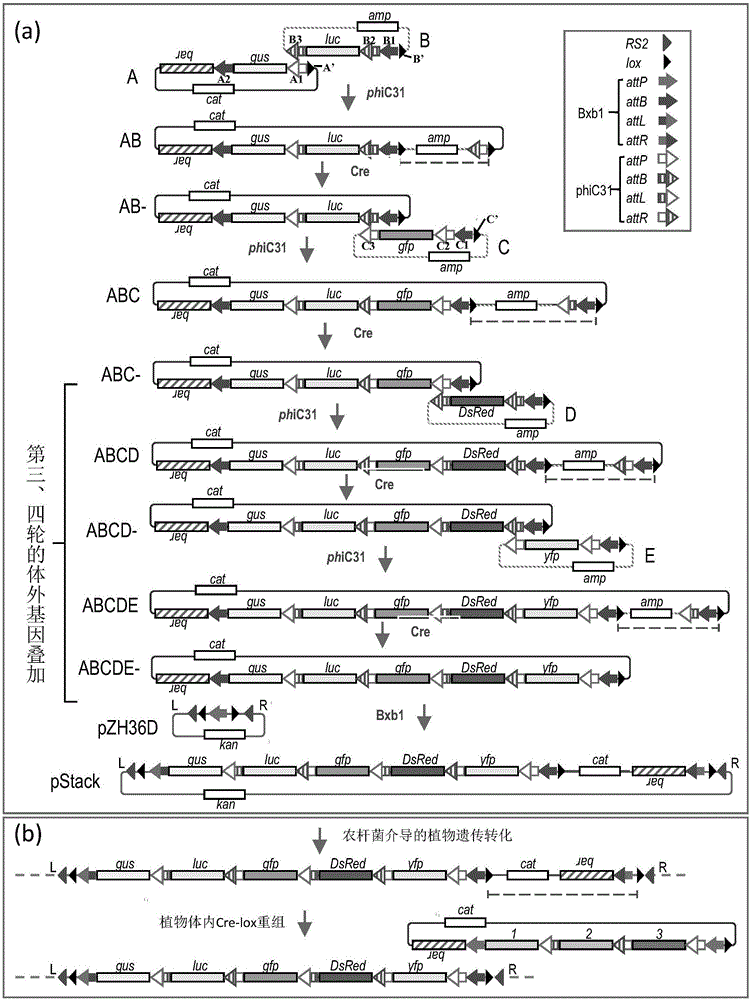
[0078] Example 1 In vitro gene stacking system
[0079] The in vitro gene stacking system includes vectors with three substrates, namely, vectors A, B, and C, and the construction process is as follows.
[0080] The conventional recombinant DNA method was used for the construction of the vector in this example. All PCR reactions used high-fidelity Phusion High-Fidelity DNA Polymerase (NEB Beijing, China). Hereinafter, the vector pZH36D uses the replication origin site of plasmid ColE1 and the replication origin site derived from plasmid pVS1 to initiate replication in Escherichia coli and Agrobacterium respectively; the rest of the vectors below include molecules A, B, C, Both D and E use the replication origin site derived from plasmid pBR322 to initiate replication. Escherichia coli DH5α ( F - endA1glnV44thi-1recA1relA1gyrA96deoRnupGΦ80dlacZΔM15Δ(lacZYA-argF)U169hsdR17(r K - m K + ), λ– ) are applied to the transformation and recovery of recombinant molecules....
Embodiment 2
[0103] The preparation of the crude extract of embodiment 2 recombinase and the conditions of in vitro recombination reaction
[0104] 1. Preparation of recombinant enzyme crude extract
[0105] Escherichia coli BL21 (Studier, F.W.andMoffatt, B.A. (1986) Useofbacteriophage-T7RNA-polymerasetodirectselectivehigh-levelexpressionofclonedgenes. J. Mol. Biol. 189 ,113-130) were selected as the prokaryotic expression host of the recombinase. Escherichia coli BL21 containing the recombinant enzyme (phiC31, Bxb1 or Cre) expression vector was first cultured at 37 degrees Celsius overnight, and then diluted 1:100 into 50ml of fresh BL medium containing 50μg / ml kanamycin, and then in Culture at 15°C to OD 600 Reach between 0.3~0.4. Then, IPTG (isopropylthiogalactopyranoside) was added to the bacterial solution to a final concentration of 0.1 mM. Continue to culture the bacterial solution at 15°C for 20 hours and then collect the bacterial cells by centrifugation (4000 rpm, 30 mi...
Embodiment 3
[0108] Example 3 Application of In Vitro Gene Overlay System
[0109] In this example, 5 exogenous target genes ( DsRed , YFP , GUS , LUC , GFP ) and transfer them into plants as an example, the superposition system described in Example 1 is used for gene superposition, and the specific operation process is as follows.
[0110] 1. The first round of in vitro gene stacking
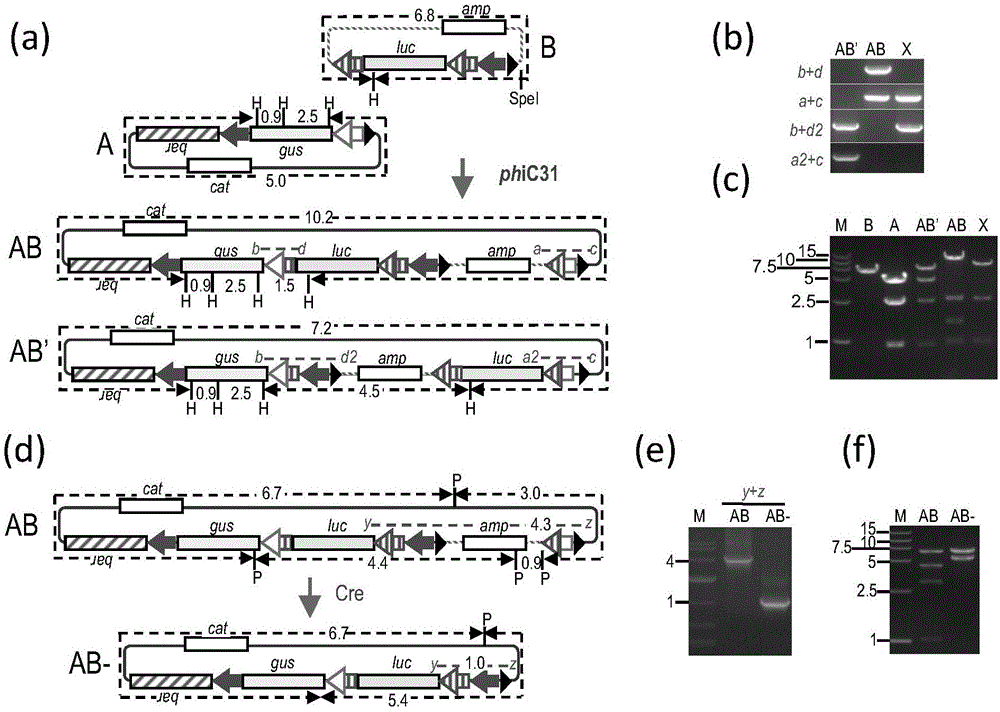
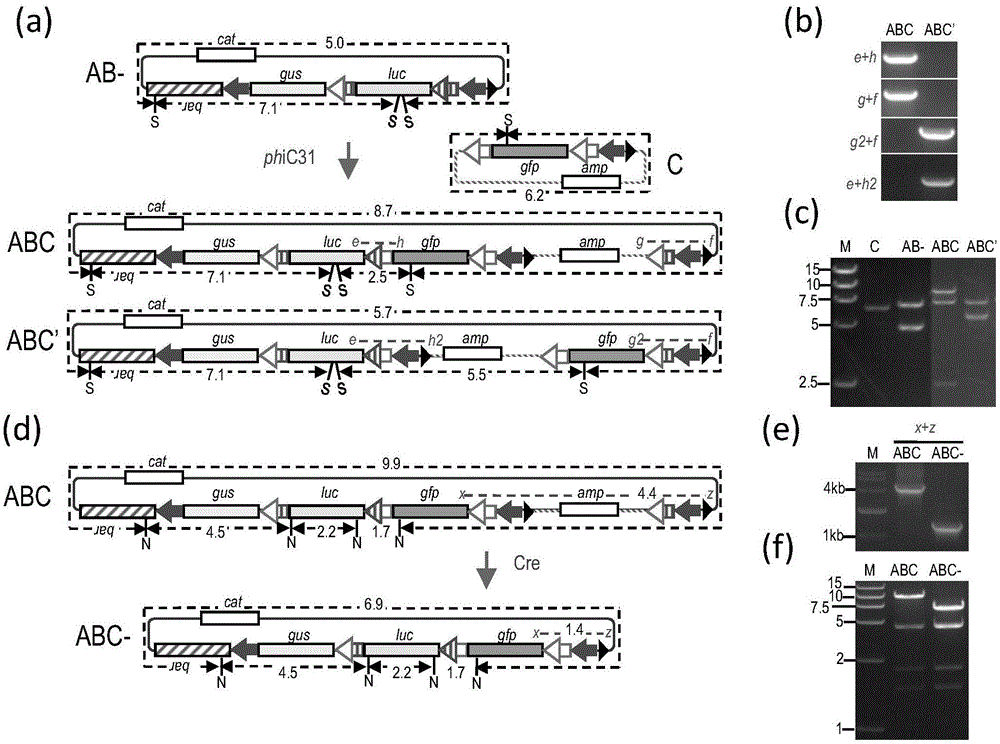
[0111] Such as figure 1 and figure 2 As shown in a, the phiC31 recombinase extract was first used to catalyze the recombination between molecules A and B (recombination products include AB and AB'), and then the recombinant DNA was recovered and transformed into E. coli DH5α. Selection was performed on solid LB medium containing chloramphenicol and ampicillin. Among the 20 clones resistant to both chloramphenicol and ampicillin, primers b+d and a+c detected 4 clones that were consistent with the molecular AB configuration, and primers b+d2 and a2+c detected 2 clones that were consistent w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com