Colon cancer marker and application thereof
A marker, colorectal cancer technology, applied in the field of biomarkers, can solve the problem of lack of non-invasive early diagnosis methods for colorectal cancer, and achieve the effect of improving and restoring the intestinal microecological structure and preventing defects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
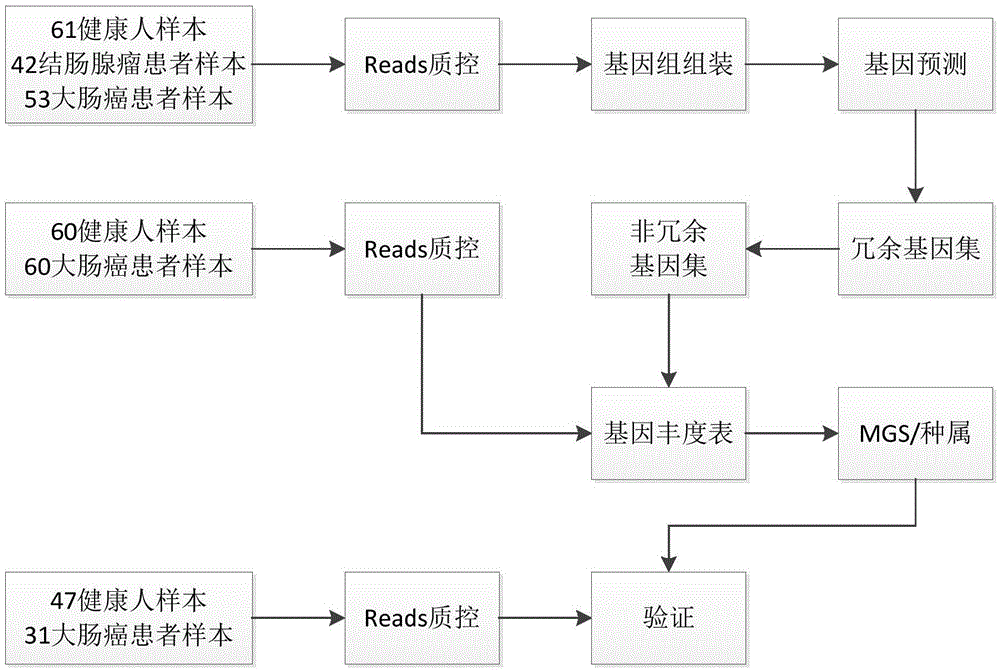
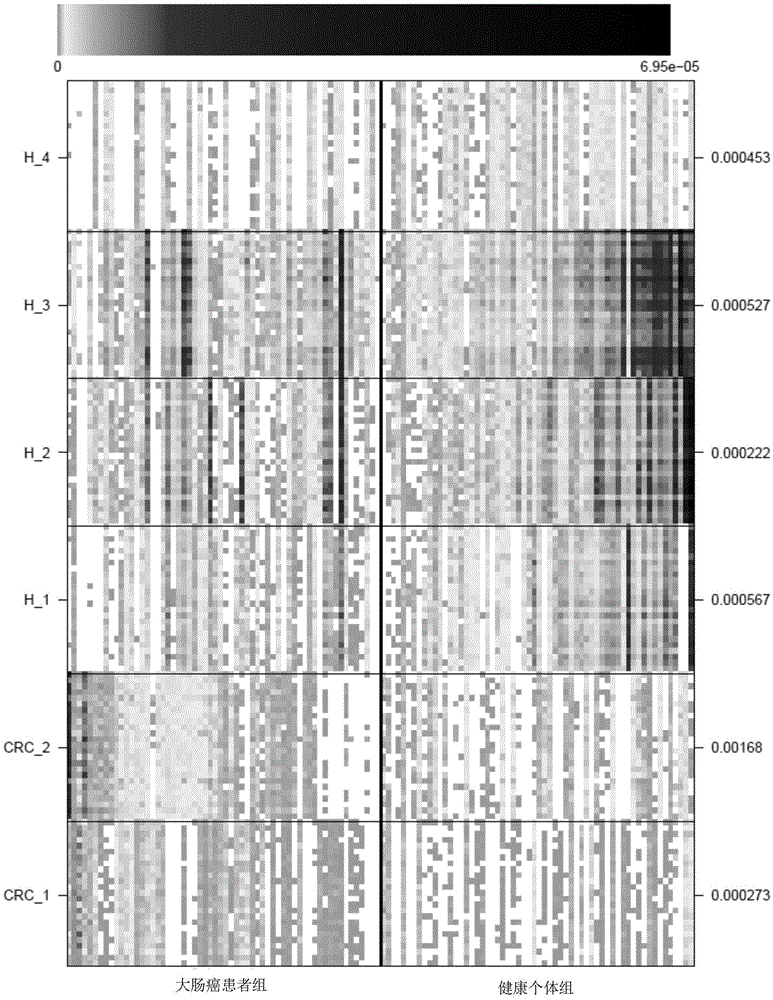
[0058] In this example, the inventors carried out correlation analysis of intestinal flora microorganisms to study the characteristics of fecal microbial communities and functional components from fecal samples of 53 patients with colorectal cancer, 42 patients with colonic adenoma, and 61 healthy individuals. In general, a total of about 1084.87Gb of sequencing data of each sample in the above-mentioned control group was downloaded, and a colorectal cancer reference gene set was constructed. Quantitative metagenomic analysis revealed that 14,993 genes were significantly different (fdr<0.05) in a large number of patients and healthy controls. Most of the genes can be classified into 6 gene clusters (MetagenomicSpecies, MGS) representing bacterial species, of which 2 MGS were enriched in the colorectal cancer patient group, and 4 MGS were enriched in the healthy group.
[0059] 1. Download the sequencing data of the control group
[0060] The DNA sequencing data of stool sampl...
Embodiment 2
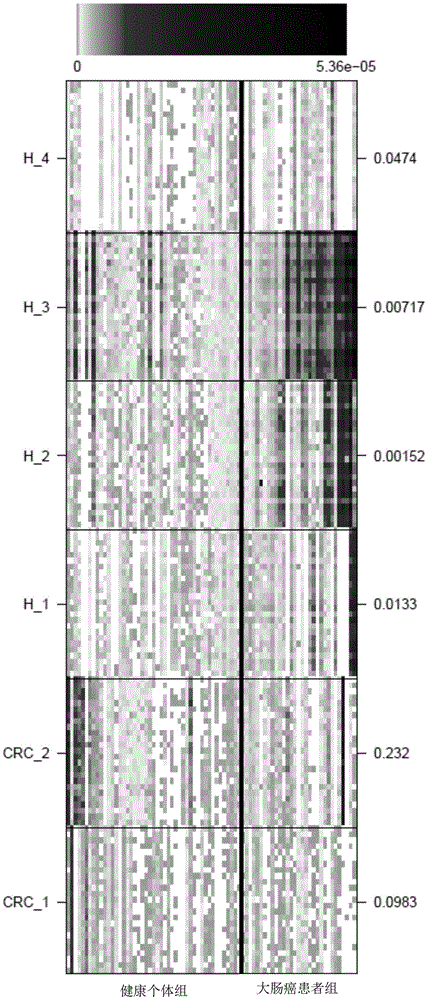
[0091] In order to verify that the species markers determined in Example 1 can be used as colorectal cancer markers, the MGS abundances of the remaining 47 healthy people and 31 colorectal cancer patients were used for verification. Wherein, the determination of gene abundance and species (MGS) abundance refers to the steps in the above examples.
[0092] The verification result is as image 3 shown. Taking the significance level α=0.05, the 4 markers determined in Example 1 that are significantly enriched in the healthy population have a significant difference in the abundance of the healthy group and the disease group in the verification group (P<0.05), as shown in Table 3. Eubacterium halllii is Enterobacter hallii, and it has been reported that enterobacterium hallii in the termite gut has a decomposing effect on lignin. Ruminococcus belongs to the genus Ruminococcus. Clostridium is the genus Clostridium and Eubacterium is the genus Eubacterium.
[0093] Taking the si...
Embodiment 3
[0097] In order to further verify whether the species markers determined in Example 1 can be used as colorectal cancer markers, stool samples from 54 colorectal cancer patients and 66 healthy individuals were obtained. The extraction of DNA in stool samples and the acquisition of sequencing data were carried out according to the instructions of the commercially available DNA extraction kits purchased and the instructions for library construction and machine use of the selected sequencing platform. The determination of gene abundance and species abundance is carried out with reference to the above examples.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com