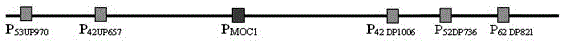Screening method of genome BAC homologous region of 12 rice species
A screening method, genome technology, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] 1 Southern probe preparation.
[0036] 1.1 Probe type.
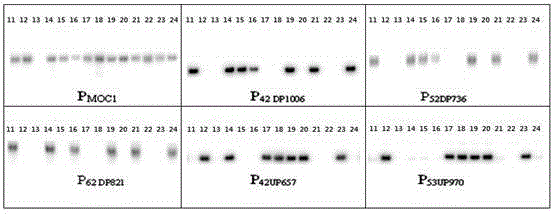
[0037]There are two types, one is "anchor probe", which is used for the first round of screening of the wild rice BAC library; the other is "limited probe", which is used for further screening of positive clones identified in the first round of screening. tiller control gene MOC1 , the latter being MOC1 Upstream 2 genes (distance from MOC1 about 42kb, 53kb respectively) probes and 3 downstream genes (distance from MOC1 about 42kb, 52kb, and 62kb respectively) probes.
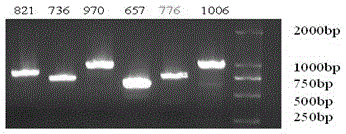
[0038] 1.2 Probe PCR amplification.
[0039] Take the japonica rice Nipponbare ( Oryza sativa L. ssp. japonica cv.Nipponbare) Genomic DNA was used as template for PCR amplification. The primer sequences are listed in Table 1.
[0040] "Anchor Probe" MOC1 Because of its high GC content (>75%), GC buffer (GCbufferⅠ, containing 5mmol / LMg2+) was used for amplification. The system is: LA Taq Enzyme, 0.3μL; 2×GCbufferⅠ, 12.5μL; dNTP, 3μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com