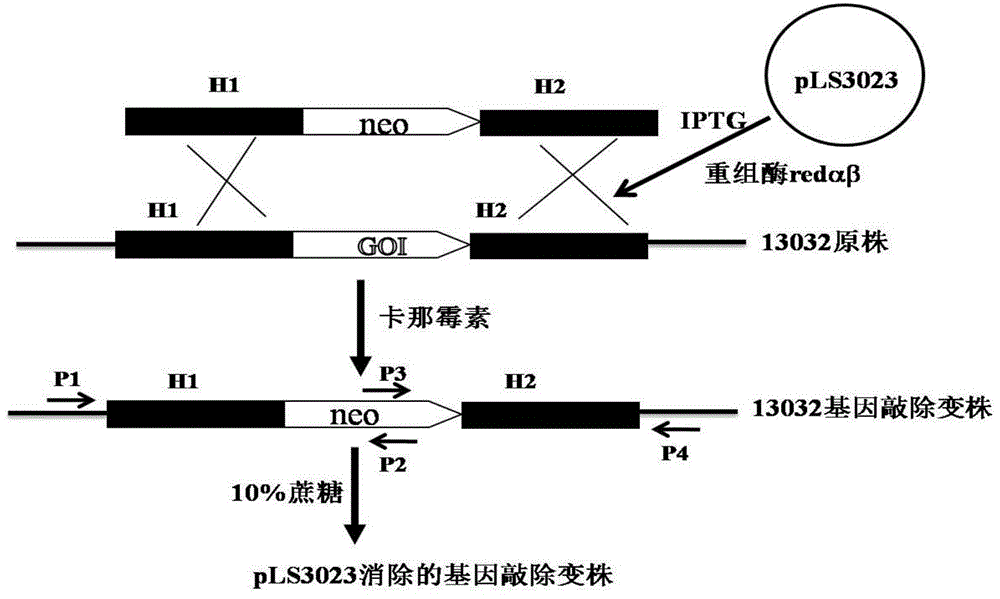
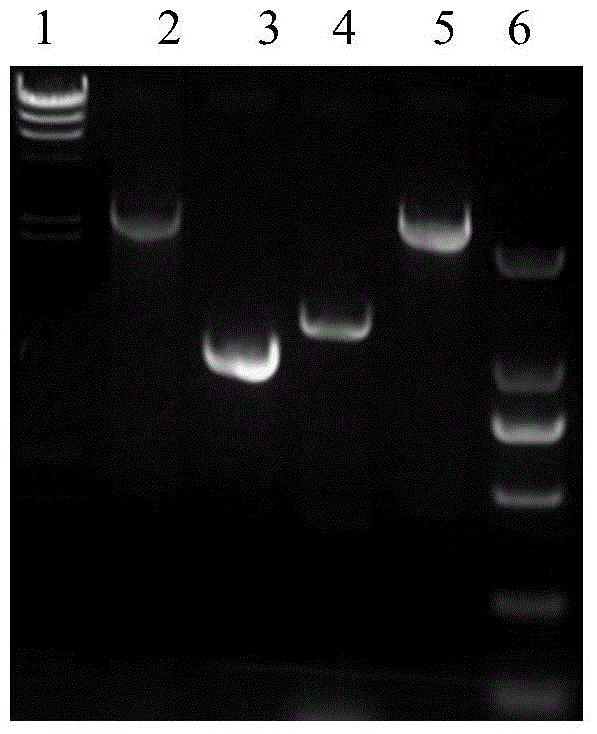
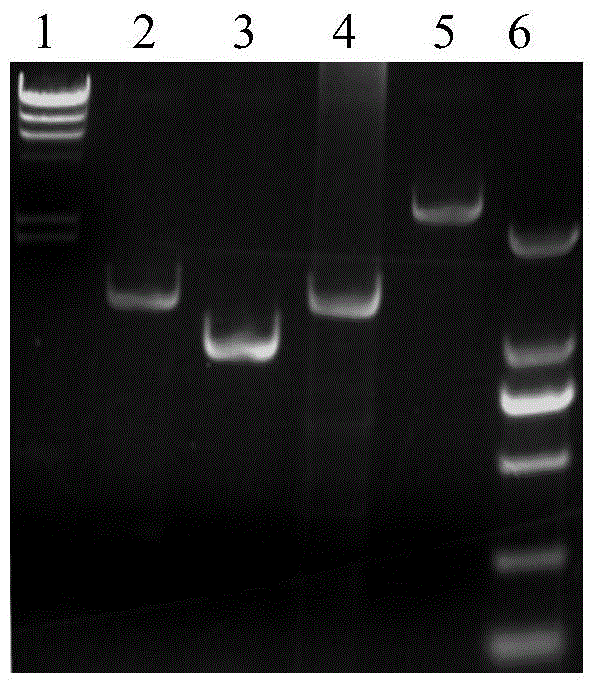
Recombineering-mediated gene knockout method of corynebacterium glutamicum ATCC 13032
A technology of ATCC13032 and Corynebacterium glutamicum, which is applied in the field of genetic engineering, can solve the problems of time-consuming, laborious, tedious, and increasing difficulty, and achieve the effect of simple and fast method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1. Construction of a plasmid that induces expression of the recombinase gene
[0044] A stuffer fragment containing the ribosome binding site is first cloned. Design primer R1407: 5'-GGG CTGCAG AAGGAGATATAGAT CCATGG CCTTCACCAGCACCTTGGTG-3', (SEQ ID NO.1), restriction sites PstI and NcoI are underlined, and the sequence between PstI and NcoI in italics is the ribosome binding site sequence, which is the gene cloned on the plasmid Required for expression in Corynebacterium glutamicum ATCC 13032; R1408: 5'-GGG GAATTC ACCCCCGGCCAGGCCAACTAC-3', (SEQ ID NO.2), the restriction site BamH is underlined. Using pSN101 as a template, R1428 and R1409 PCR to obtain a 1.5kb fragment, 1.5kb was digested with PstI and BamHI, and 3.0kb was ligated with pBluescript II KS(-) digested with the same enzyme, and transformed into Escherichia coli (Escherichia coli) DH10B. Transform competent cells in the presence of isopropyl-β-D-thiogalactoside (IPTG) and 5-bromo-4-chloro-3-ind...
Embodiment 2
[0047] Example 2. Knockout of the crtI2 gene on the genome of Corynebacterium glutamicum ATCC 13032
[0048] 1. Preparation of Corynebacterium glutamicum ATCC 13032 electroporation competent cells and transformation of pLS2406
[0049] Corynebacterium glutamicum ATCC 13032 was streaked from a -80°C cryopreservation tube to BHIS solid medium, and cultured at 30°C for 24 hours. Transfer a single colony to 3ml BHIS liquid medium, culture at 30°C and 220rpm for about 24 hours, then transfer 2ml to 100ml BHIS, adjust the OD600 to about ~0.1, continue to cultivate until the OD600 is about 0.6, centrifuge, and discard the supernatant. The cell pellet was gently washed three times with ice-cold 10% glycerol, and finally suspended in 200 μl of ice-cold 10% glycerol, and each 50 μl was dispensed into an Eppendorf tube. Add 100ng of pLS2303 plasmid DNA to the competent cells, flick and mix well, transfer to a 2mm electric cuvette cooled on ice, and transform with 2.5kV electric shock. T...
Embodiment 3
[0059] Example 3. Knockout of the upp gene on the genome of Corynebacterium glutamicum ATCC 13032
[0060] Three pairs of primers R1431 were designed according to the sequence of the upp gene on the genome of Corynebacterium glutamicum ATCC 13032 and the sequence of the kanamycin resistance gene: 5'-ATAAAAGGGTGGTGGGGTTGGC-3', (SEQ ID NO.15), R1432: 5' - GCGCGGAACCCCTATTTGTTCATAGCTTCACATGTTAAATCATTGC-3', (SEQ ID NO. 16), R1433: 5'-GCAATGATTTAACATGTGAAGCTATGAACAAATAGGGGTTCCGCGC-3', (SEQ ID NO. 17), R1434: 5'-CCGTAATGCCCTTAGAAACTTTTATCCTTAGTTCCTATTCCGAAG, IDNO. R1435: 5'-CTTCGGAATAGGAACTAAGGATAAAAGTTTCTAAGGGCATTACGG-3', (SEQ ID NO. 19), R1436: 5'-GGAGGATGAAGTCCAAAGCTG-3', (SEQ ID NO. 20). Among them, R1432 and R1433, R1434 and R1435 are all reverse complementary to each other, and are used for amplification of overlap-extension PCR.
[0061] Using Corynebacterium glutamicum ATCC 13032 genomic DNA as a template, R1431 and R1432 PCR amplified to obtain a fragment of 500 bp upstrea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com