Fusion nucleic acid, fusion protein, and kit for detecting secondary glioblastoma
一种胶质母细胞瘤、融合蛋白的技术,应用在检测继发胶质母细胞瘤的试剂盒领域,能够解决继发性胶质母细胞瘤和原发性胶质母细胞瘤准确性有待提高等问题,达到提高准确性的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation Embodiment 1
[0051] In this preparation example, secondary glioblastoma samples and primary glioblastoma samples were obtained, and their RNA and cDNA were obtained.
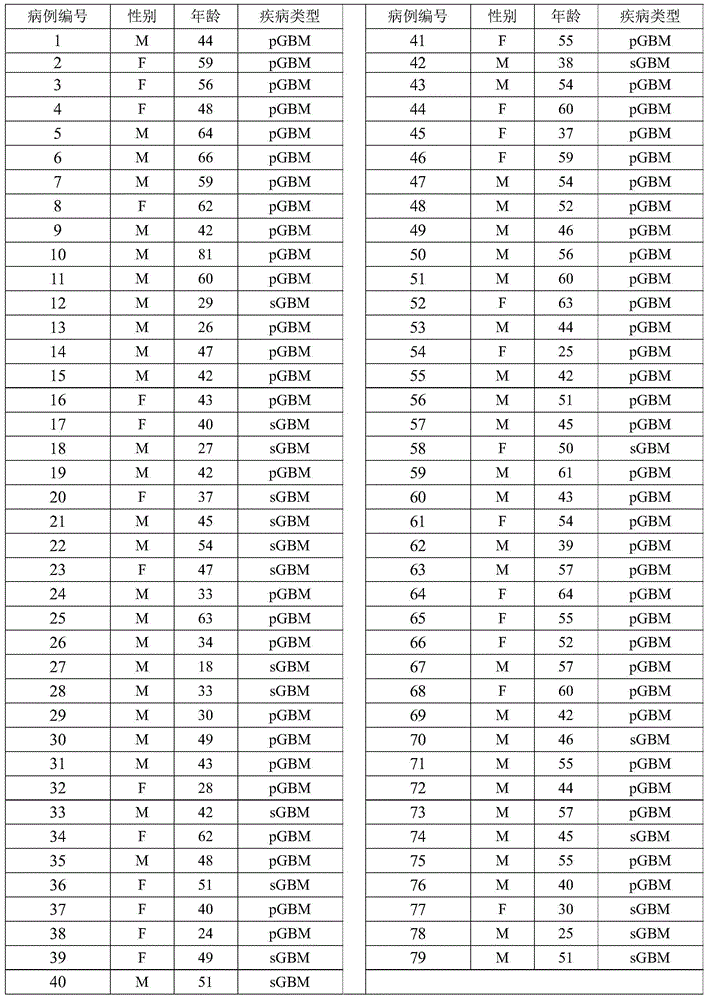
[0052] Table 1
[0053]
[0054] 59 samples of primary glioblastoma (primary glioblastoma) and 20 samples of secondary glioblastoma (secondary glioblastoma) were collected using the operation in accordance with the standards of the medical ethics committee. Among them, each patient who collects samples has obtained the consent of himself and his therapist before collecting samples, and has written certification materials. Among them, the diagnosis and differentiation of primary glioblastoma and secondary glioblastoma are based on the histology in the literature (LouisDN, et al, 2007. The2007WHOclassificationoftumoursthecentralnervoussystem.ActaNeuropathol114(2):97-109.) method carried out. The gender, age, and disease type information of the pathological samples are shown in Table 1, where pGBM represents primary gliobl...
Embodiment 1
[0057] In this embodiment, the RNA of 59 primary glioblastoma samples and 20 secondary glioblastoma samples collected in Preparation Example 1 were sequenced.
[0058] The RNA of each sample was constructed into an RNA library using an RNA library construction kit (purchased from Illumina), and then RNA sequencing was performed on the RNA library using a sequencing platform (Illumina HiSeq2000). The sequence obtained by sequencing was compared with the reference RNA sequence database (Hg19Refseq, GRCh37), and the method in the reference (McPhersonA, etal.
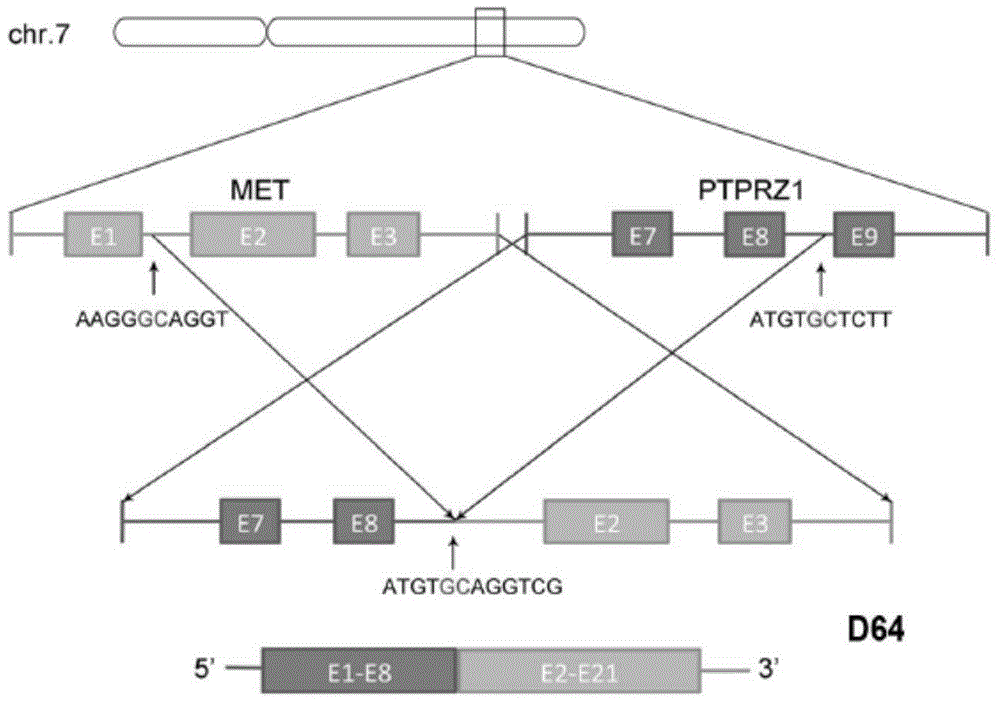
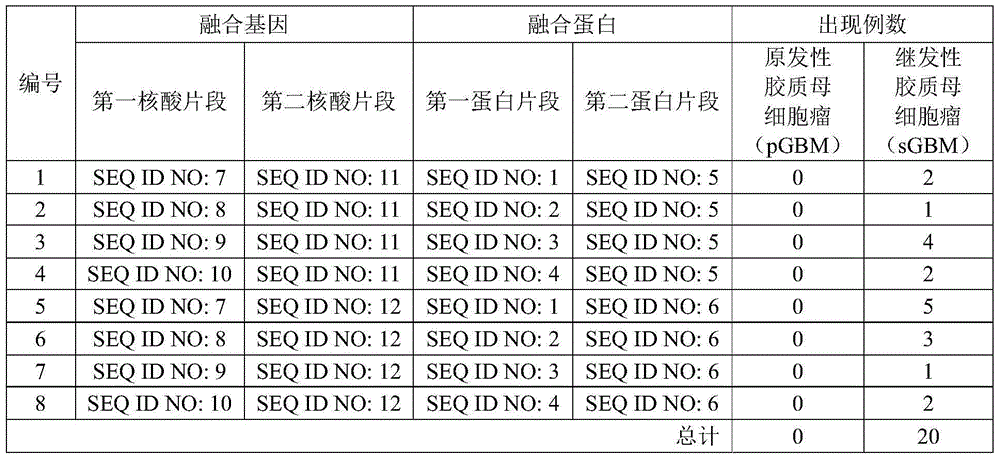
[0059] As a result, it was found that in the samples shown in Table 1, the RNA of the fusion gene of the present invention appeared in the samples of multiple secondary glioblastomas (sGBM), but the primary glioblastoma (pGBM) No RNA of the fusion gene of the present invention appeared in any of the samples, and the specific occurrences are shown in Table 2.
[0060] Table 2
[0061]
[0062] It can be seen from the da...
Embodiment 2
[0065] In this embodiment, PCR verification of the fusion protein was carried out on the cDNA obtained from RNA prepared from 59 samples of primary glioblastoma and 20 samples of secondary glioblastoma collected in Example 1.
[0066] The primers used for PCR verification consist of the first primer shown in SEQ ID NO:17 and the second primer shown in SEQ ID NO:18. The operation of PCR was carried out according to the instructions of synthetic primers and PCR kit. The product of PCR displays the presence or absence of amplified bands through agarose gel nucleic acid electrophoresis, and the amplified bands that appear are recovered using a DNA gel recovery kit (QIAquickPCR purification kit, purchased from Qiagen), and then cloned into a T vector (pGEM - Teasyvector, purchased from Promega), and sequenced with a DNA sequencer (ABIPrism3730×1 DNA Sequencer, purchased from Applied Biosystems). The results are shown in Table 3
[0067] table 3
[0068]
[0069] It can be see...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com