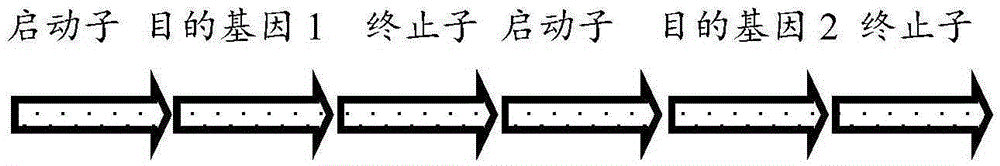
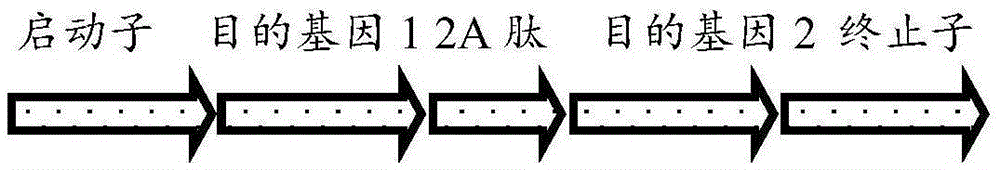
Microalgae multigene co-expression vector and multigene co-expression microalgae
A technology of co-expression vector and expression vector, applied in the field of genetic engineering of microalgae
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Construction of pEASY-F2A, P2A, T2A recombinant plasmids
[0030]According to the amino acid sequences of the three 2A peptides of F2A, P2A, and T2A and the endogenous codon preference of Chlorella pyrenoidosa[8], the nucleotide sequences were designed, and two upstream and downstream primers with a repeat of about 20bp were designed, respectively. For F2A1, F2A2, P2A1, P2A2, T2A1, T2A2, the complete sequence was synthesized by the fusion PCR method, recovered and purified, and then connected to the pEASY-Blunt cloning vector, using M13 primers for PCR to identify single colonies, and sequencing verification after extracting the plasmid. The primers used are as follows:
[0031]
[0032]
[0033] Primers used during the fusion of table 12A peptides
Embodiment 2
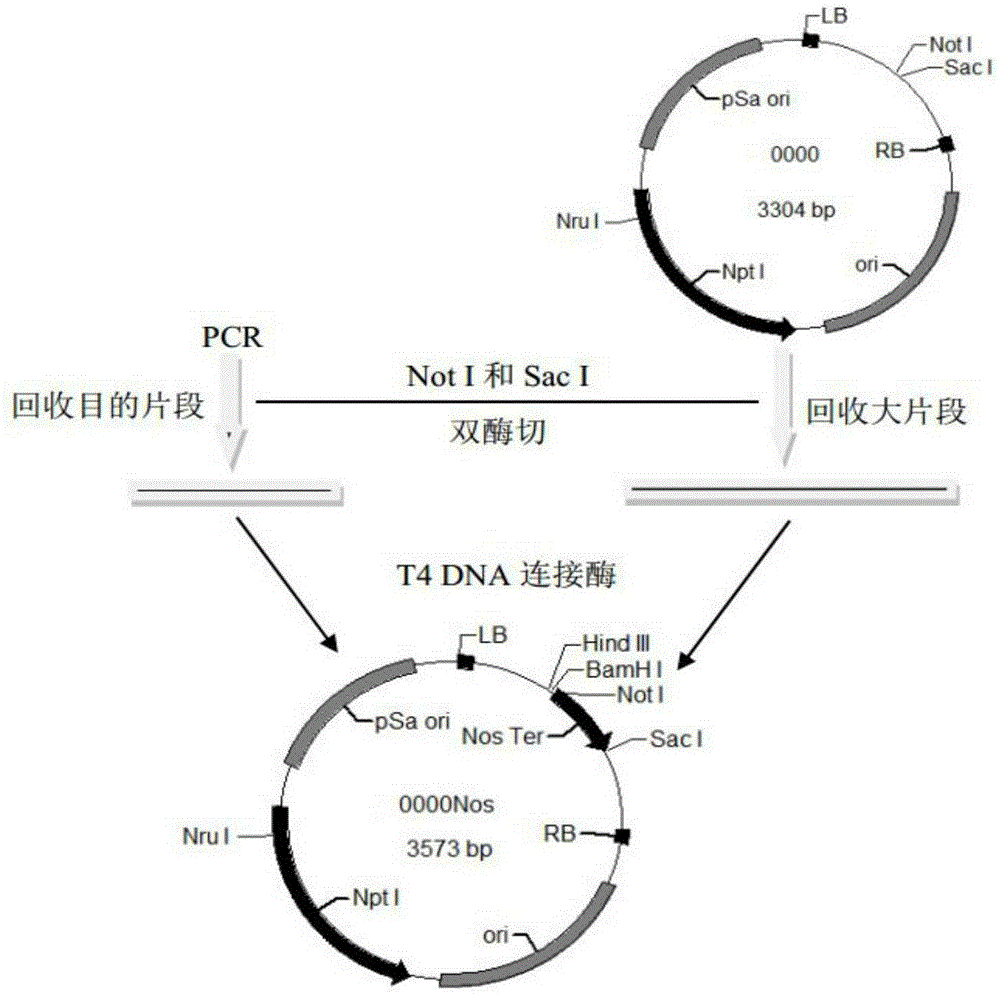
[0035] Construction of pGreenII0000-NosT recombinant plasmid
[0036] refer to image 3 As shown, the Nos terminator fragment was cloned from the pGreenII0029 plasmid with Nos terminator upstream and downstream primers NosT-F / R containing NotI and SacI restriction sites respectively and high-fidelity enzyme Pfu polymerase. Digest the pGreenII0000 plasmid and the Nos terminator fragment with SpeI and NotI, isolate and purify the Nos terminator fragment and the linearized pGreenII0000 plasmid, and then use T4 DNA ligase to ligate overnight to obtain the pGreenII0000-NosT recombinant plasmid, which is transformed into Escherichia coli Trans-T1 State cells, use 00F and 00R primers to do PCR to identify single colonies, and extract plasmids for sequencing verification. The primers used are as follows:
[0037]
[0038] Primers used when table 2 constructs pGreenII0000-NosT recombinant plasmid
Embodiment 3
[0040] Construction of pGreenII0000-NptII-P2A-EGFP-NosT recombinant plasmid
[0041] refer to Figure 4 As shown, first use NptII respectively Bam F and NptII P2A R primer cloned NptII gene fragment (neomycin phosphotransferase), P2A NptII F and P2A EGFP R primer cloned P2A fragment, EGFP P2A F and EGFP Not The R primer clones the EGFP fragment, where NptII P2A R and P2A NptII F primer is reverse complementary sequence, containing 18bp NptII gene end sequence and 18bp P2A gene front sequence, P2A EGFP R and EGFP P2A The F primer is a reverse complementary sequence, containing 18 bp of the end sequence of the P2A gene, 19 bp of the front end sequence of the EGFP gene and the sequence of the SpeI restriction site. After recovering and purifying the three gene fragments, connect the three fragments by recombinant PCR method to obtain the NptII-P2A-EGFP sequence, and then use NptI IBam F and EGFP Not R primers amplify this sequence, recover and purify it and connect it ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com