A method for rapid quantitative determination of microbial community composition during liquor fermentation
A microbial community and fermentation process technology, applied in the field of rapid PSP-qPCR quantification, can solve the problems of high cost, inability to meet the composition and structure of microbial community, and high quality requirements of endonucleases, so as to achieve low cost, improve wine production efficiency and effect significant effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
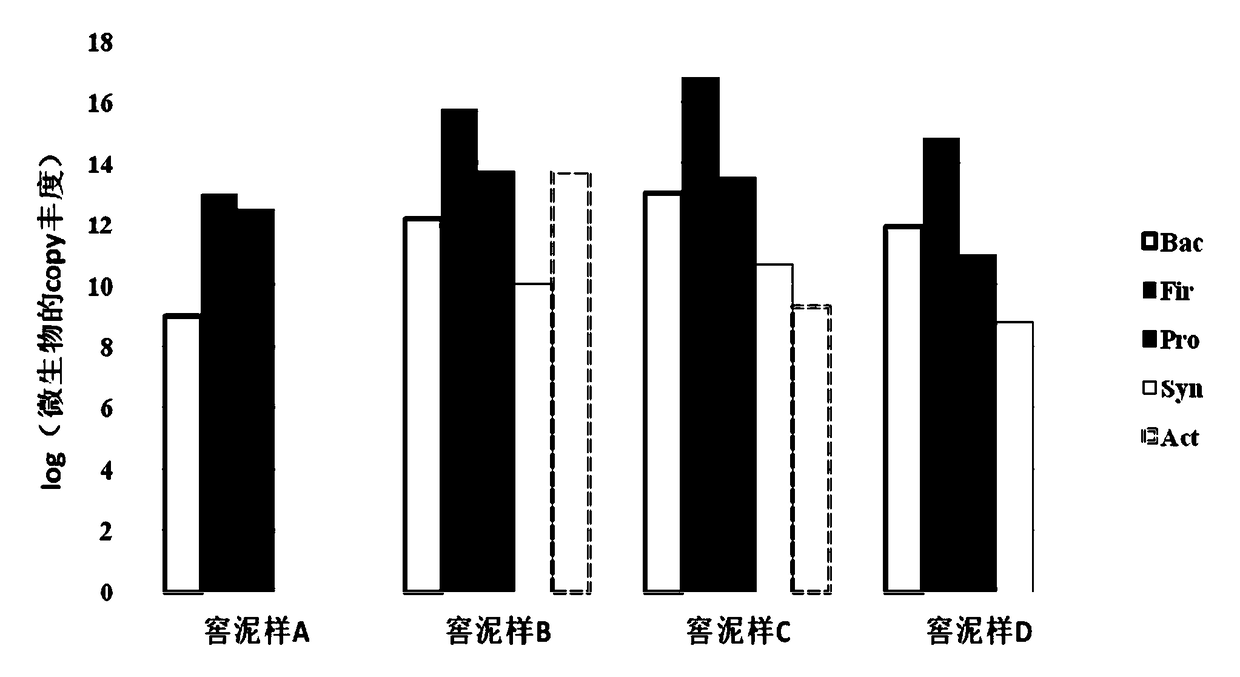
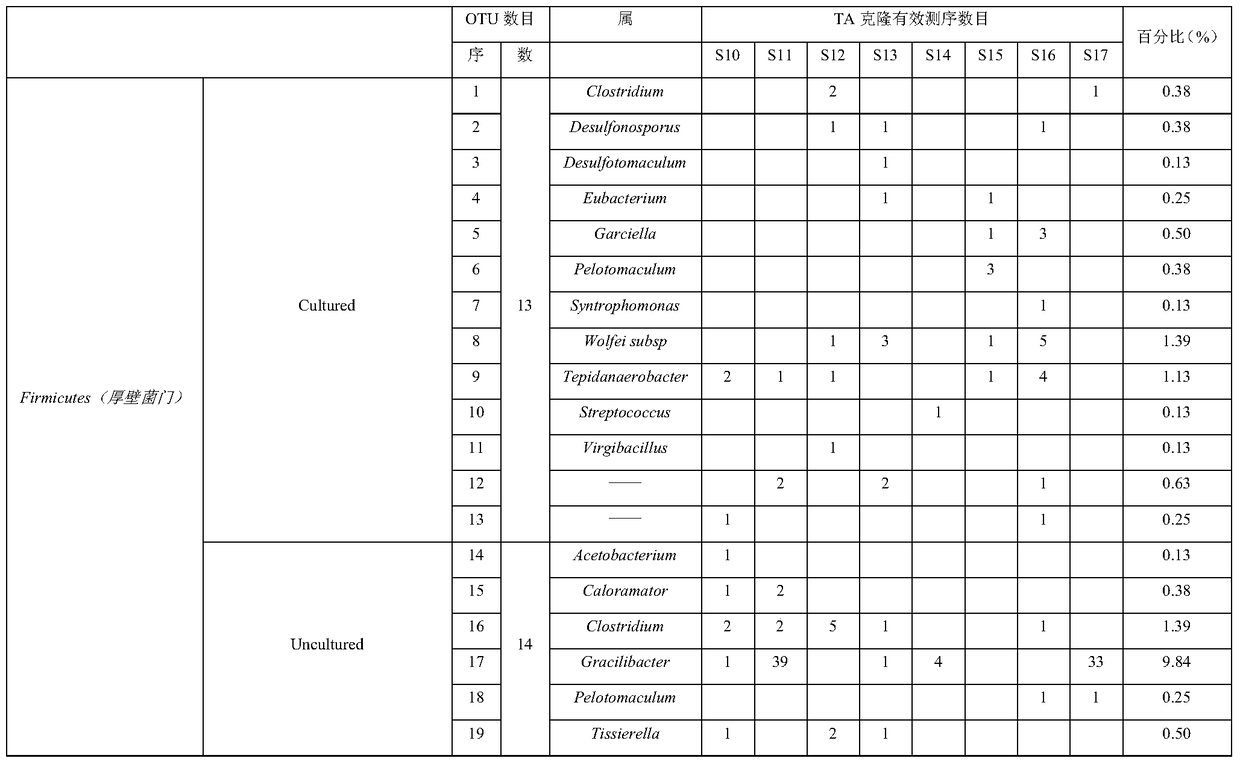
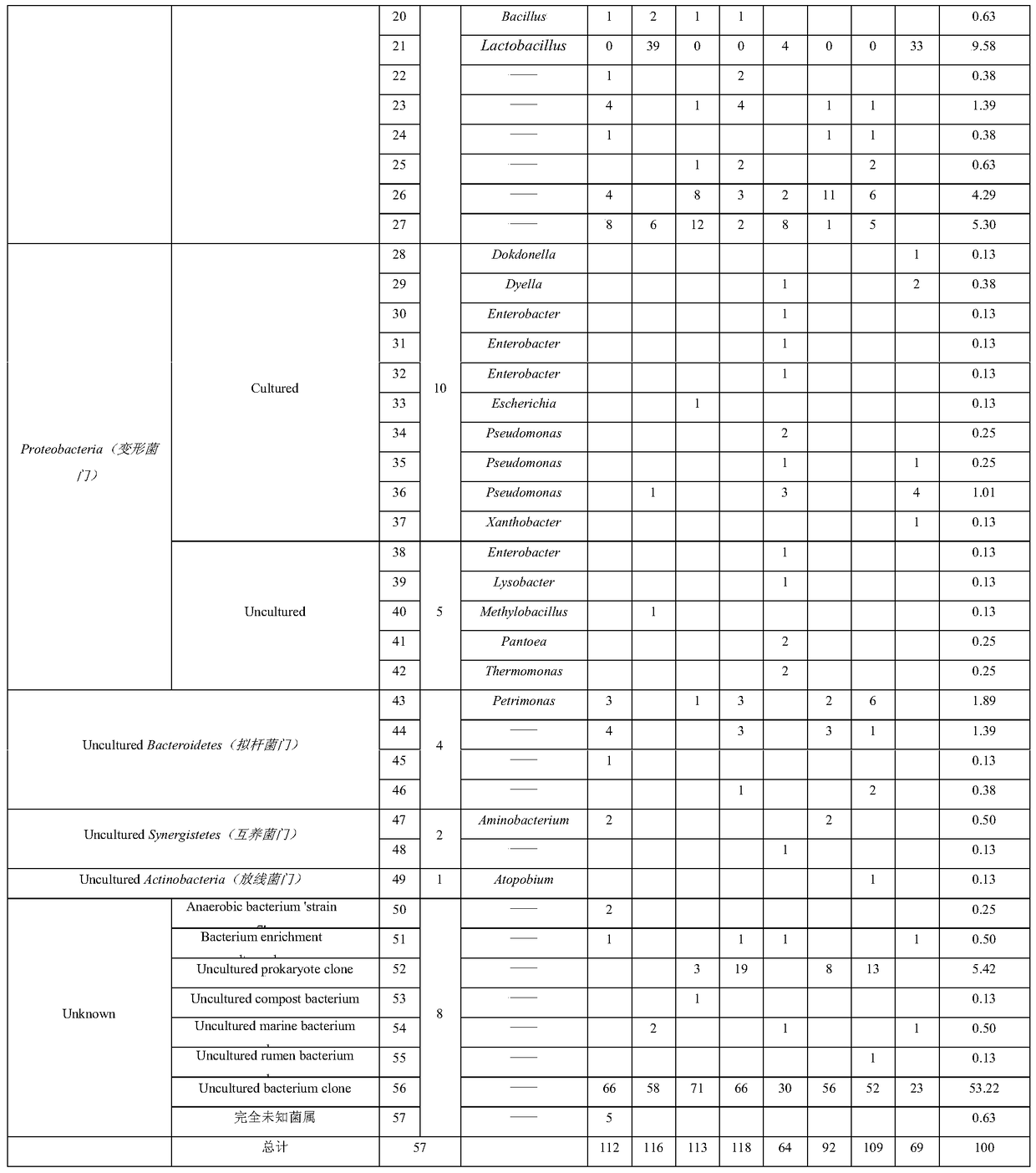
[0043] In this example, the method for rapidly determining the composition of the microbial community in the liquor fermentation process is to use phylum-specific primers in the PCR reaction system to specifically amplify the microbial flora of a specific phylum during the liquor fermentation process, and use qPCR The system performs quantitative amplification to obtain quantitative identification.
[0044] Prepare gate-specific primers as follows:
[0045] Step a, extracting the metagenomic group of the brewing microbial community sample as a PCR amplification template;
[0046] Step b, amplifying the ribosomal DNA in the amplified template is close to the full-length 16s rDNA sequence, and the amplification primers are 27F (5'-AGA GTT TGA TCC TGG CTC AG-3') and 1492R (5'-GGT TAC CTT GTT ACG ACTT-3'), obtain the amplification product;
[0047] Step c. Perform TA cloning, blue-white screening and white-white colony bidirectional sequencing sequentially after the amplificatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com