Method for batch computing of evolutionary rate of orthologous genes of genome
A homologous gene and genome technology, applied in the field of batch calculation of the evolution rate of genome orthologous genes, can solve the difficulty of increasing the batch calculation of the evolution rate of genome orthologous genes, limit the use of non-biological information professionals, and restrict the field of molecular evolution Research and other issues to achieve the effect of making up for time-consuming and labor-intensive, fast, comprehensive and comprehensive parameters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1. Establishment of a method for calculating the evolution rate of genome orthologous genes in batches
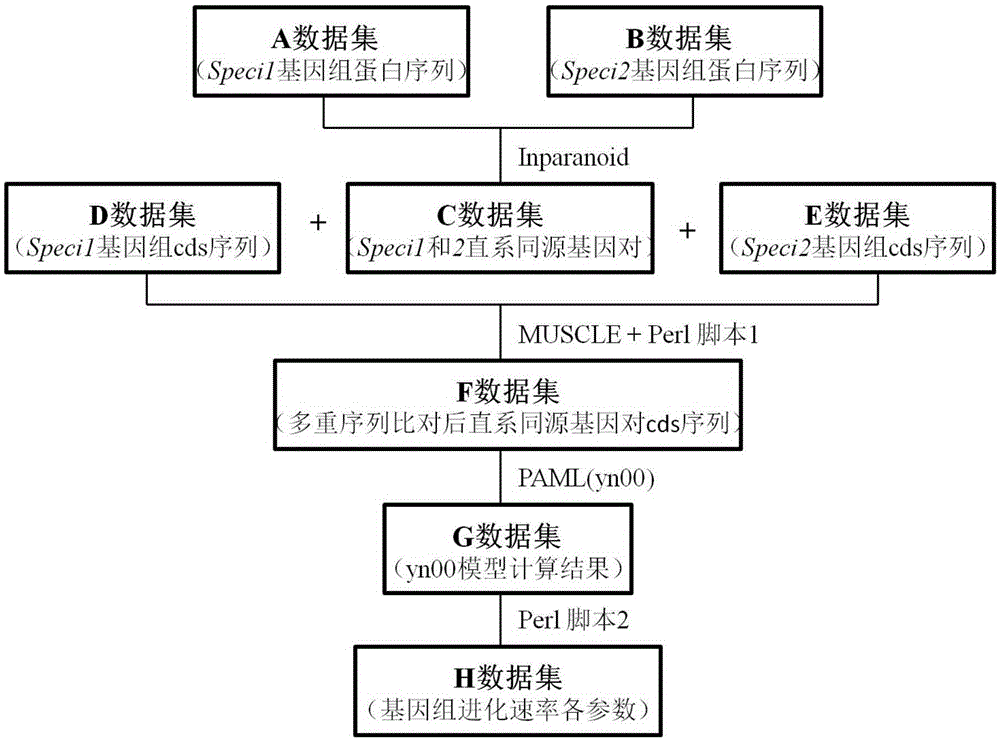
[0034] The flowchart of the method for calculating the evolution rate of genome orthologous genes in batches provided by the present invention is shown in figure 1 , including the following steps:
[0035] (1) Using the InParanoid program for clustering based on the results of Blast pairwise comparisons, identify the orthologous genes in the Speci1 and Speci2 genomes (data sets A and B) to be tested, operate under the Linux system, and use default parameter settings Perform analysis; follow the steps below to obtain complete orthologous gene pair ID and score information files (C data set);
[0036] Steps to obtain the complete orthologous gene pair ID and score information file: Open the folder where the InParanoid software package "inparanoid.pl" file is located, and run the "perlinparanoid.plXXX1XXX2" command, where "XXX1" and "XXX2" represent A , the fil...
Embodiment 2
[0055] Example 2, using the method established in Example 1 to calculate the evolution rate of genes between Raymond cotton and Asian cotton genomes in batches
[0056] Enter the cotton genome project link (http: / / cgp.genomics.org.cn / ) database of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences to download the genome sequence (13 chromosomes, 885Mb) and Asian cotton (Gossypium arboretum L.) (13 chromosomes, 1,746Mb), in the Windows system or local Linux computing server, the gene evolution rate between Raymond cotton and Asian cotton genomes was calculated. During the calculation process, the commonly used program names, operating environments and addresses involved are shown in Table 1. The specific operation steps of the calculation method are as follows:
[0057] 1) Carry out with reference to the step (1) of Example 1.
[0058] Using the InParanoid program to identify the orthologous genes between the genomes of Raymond cotton (data set A) a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com