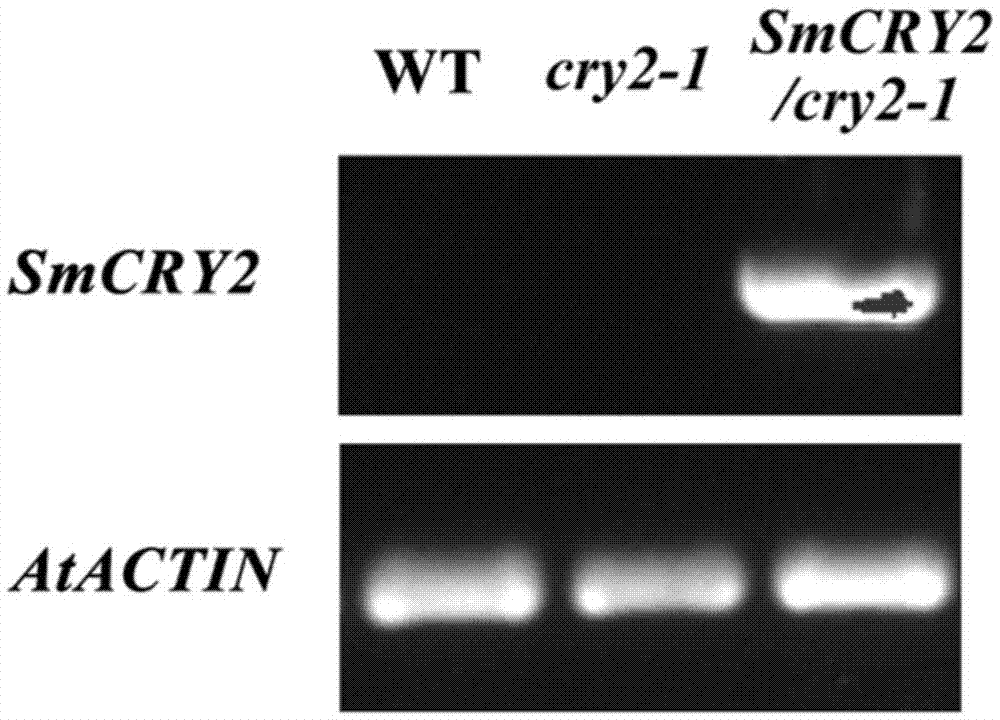
Eggplant cryptochrome gene SmCRY2 and application thereof
A gene, eggplant technology, applied in the field of eggplant cryptochrome gene SmCRY2, can solve problems such as unknown
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, the cloning of eggplant SmCRY2 gene
[0039] 1. Acquisition of plant material
[0040] The plant material used in this experiment is the excellent eggplant germplasm resource A. spp. The experimental materials were cultivated in artificial plastic greenhouses in the Aerospace Breeding Base of Pujiang Town, Minhang District, Shanghai. Seedlings are raised, grown and fruited under natural conditions. The leaves of eggplant were collected for RNA extraction.
[0041] 2. Extraction of RNA
[0042]Total RNA was extracted by the TRIzol method (TRIzol was purchased from Sangon Bioengineering (Shanghai) Co., Ltd.). The integrity of RNA was identified by formaldehyde denaturing gel electrophoresis, and then the purity and concentration of RNA were determined on a spectrophotometer (ThermoScientificNANODROP1000Spectrophotometer).
[0043] 3. Full-length cloning of genes
[0044] A pair of degenerate primers were designed based on the conserved sequence of CR...
Embodiment 2
[0048] Example 2. Sequence information and homology analysis of eggplant CRY2 gene
[0049] The full-length CDS open reading frame sequence of the new eggplant CRY2 gene of the present invention is 1944bp, and the detailed sequence is shown in SEQ ID NO.1. According to the CDS open reading frame sequence, the amino acid sequence of eggplant CRY2 is deduced, with a total of 647 amino acid residues, a molecular weight of 73208.5 Daltons, and a theoretical isoelectric point (pI) of 5.91. The detailed sequence is shown in SEQ ID NO.2.
[0050] The CDS open reading frame sequence of eggplant CRY2 and the amino acid sequence of its encoded protein were identified by BLAST program in the Non-redundantGenBank+EMBL+DDBJ+PDB and Non-redundantGenBankCDStranslations+PDB+SwissProt+Superdate+PIR databases. Source retrieval, it was found that it has 92.28% identity with the tomato CRY2 gene (NM_001247316.1) at the nucleotide level; at the amino acid level, the similarity between them is as...
Embodiment 3
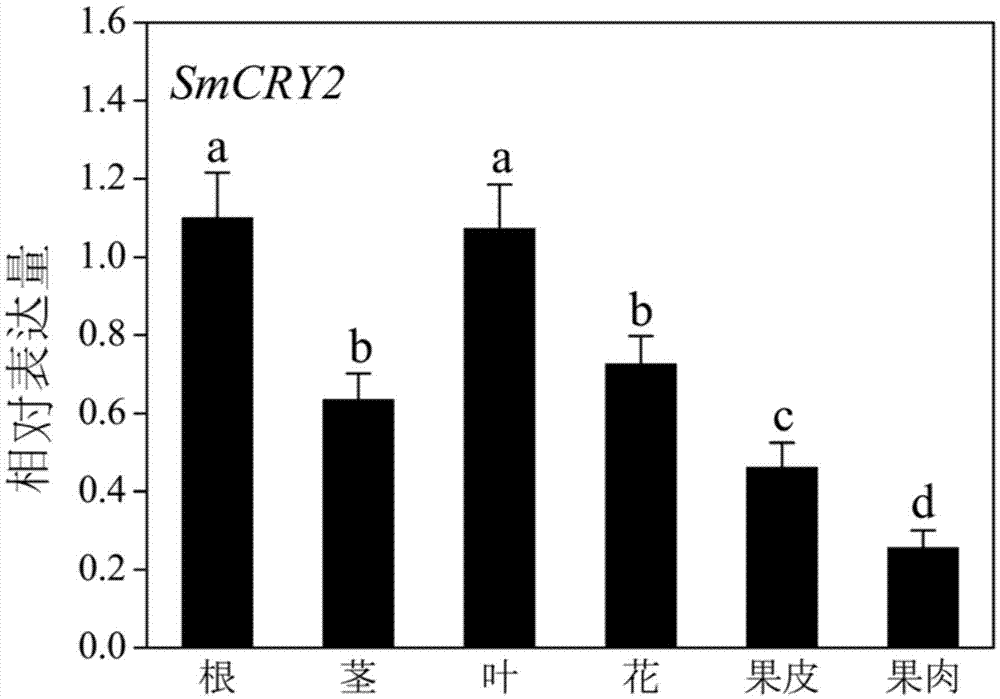
[0051] Embodiment 3, expression of eggplant CRY2 gene in different tissues
[0052] 1. Acquisition of plant material
[0053] The eggplant roots, stems, leaves, flowers, pericarp, and pulp were collected at the ripening stage of the fruit, and the samples were wrapped in aluminum platinum paper and immediately put into liquid nitrogen, and then transferred to a -80°C ultra-low temperature refrigerator for storage until use.
[0054] 2. Extraction of RNA
[0055] Total RNA was extracted by the TRIzol method (TRIzol was purchased from Sangon Bioengineering (Shanghai) Co., Ltd.). The integrity was detected by ordinary agarose gel electrophoresis (gel concentration 1.2%; 0.5×TBE electrophoresis buffer; 150v, 15min). The maximum rRNA brightness in the electrophoresis strip should be 1.5-2.0 times the brightness of the second rRNA, otherwise it indicates the degradation of the rRNA sample. RNA with good purity, A 260 / A 280 and A 260 / A 230 About 2.0 or so. The OD value wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com