Method of detecting single base mutation and application
A single-base mutation, gene technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of complex operation, high price, time-consuming, etc., and achieve the effect of good repeatability and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0053] The present invention will be described in detail below in conjunction with the embodiments and accompanying drawings, but the present invention is not limited thereto.
[0054] A method for detecting a single base mutation, comprising the steps of:
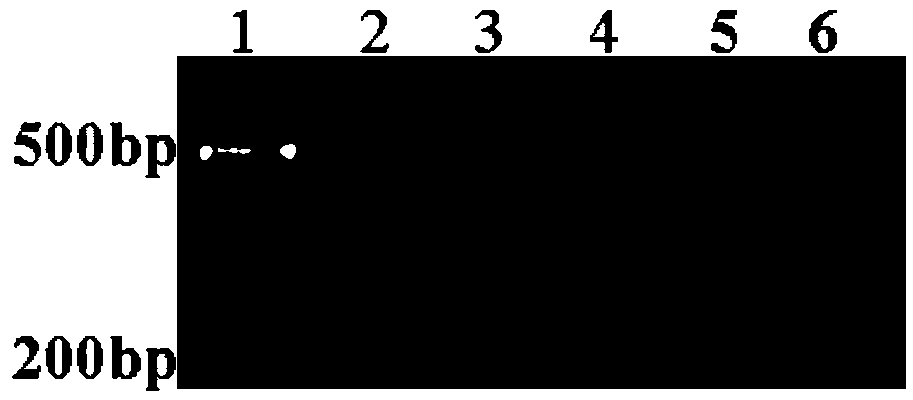
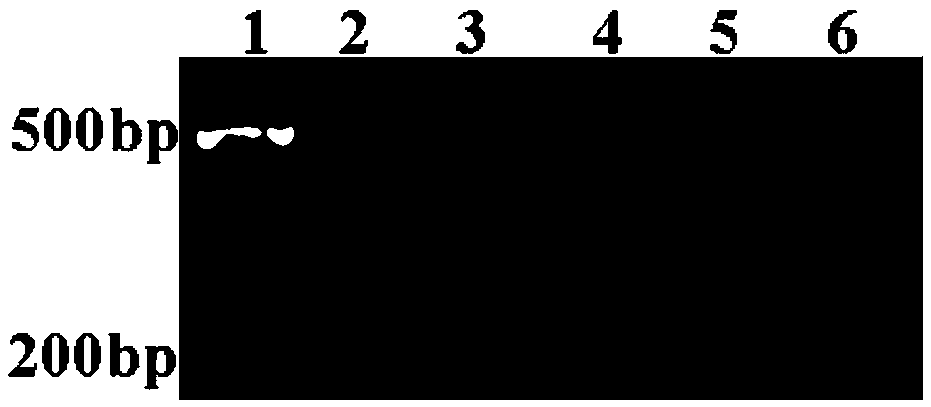
[0055] (1) PCR amplification of the target segment
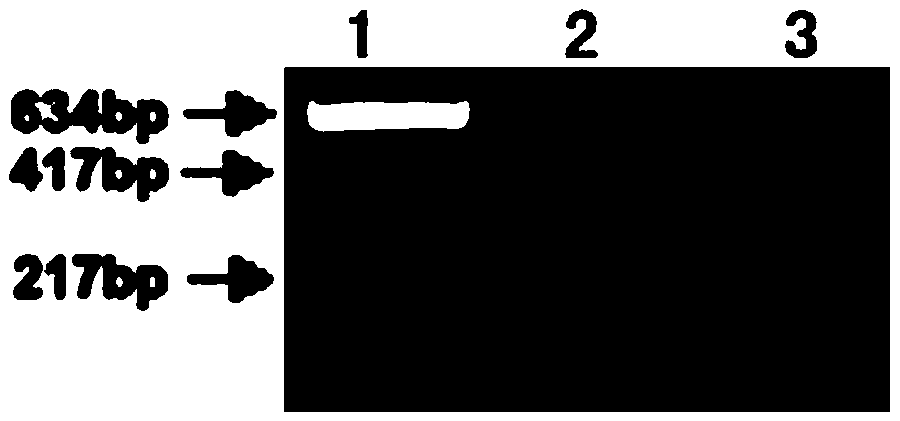
[0056] A pair of primers were designed according to the gene sequence encoding celery nuclease CELI, and PCR was used to amplify the fragment to be detected from two different celery DNA samples (CA and CB), with a size of 634bp. The upstream and downstream primers used were synthesized at Shanghai Sangon Biotechnology Co., Ltd., and the sequences are as follows:
[0057] CPF: 5'-ACACCTGATCAAGCCTGTTCATTTGATTAC-3';
[0058] CPR: 5'-CGCCAAAGAATGTACTGCGGAGCTT-3'.
[0059] The PCR reaction system is as follows:
[0060]
[0061] The reaction instrument used MG96GPCR instrument, and the amplification program was as follows: pre-denaturation at 94°C for 3 minutes; denat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com