Identification and use of circulating nucleic acid tumor markers
A tumor and oligonucleotide technology, which is applied in biochemical equipment and methods, microbial determination/inspection, bioinformatics, etc., and can solve the problems of availability and inability to detect the existence of mutations in plasma samples.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0782] Example 1: Ultra-sensitive method for quantifying circulating tumor DNA in a wide range of patients
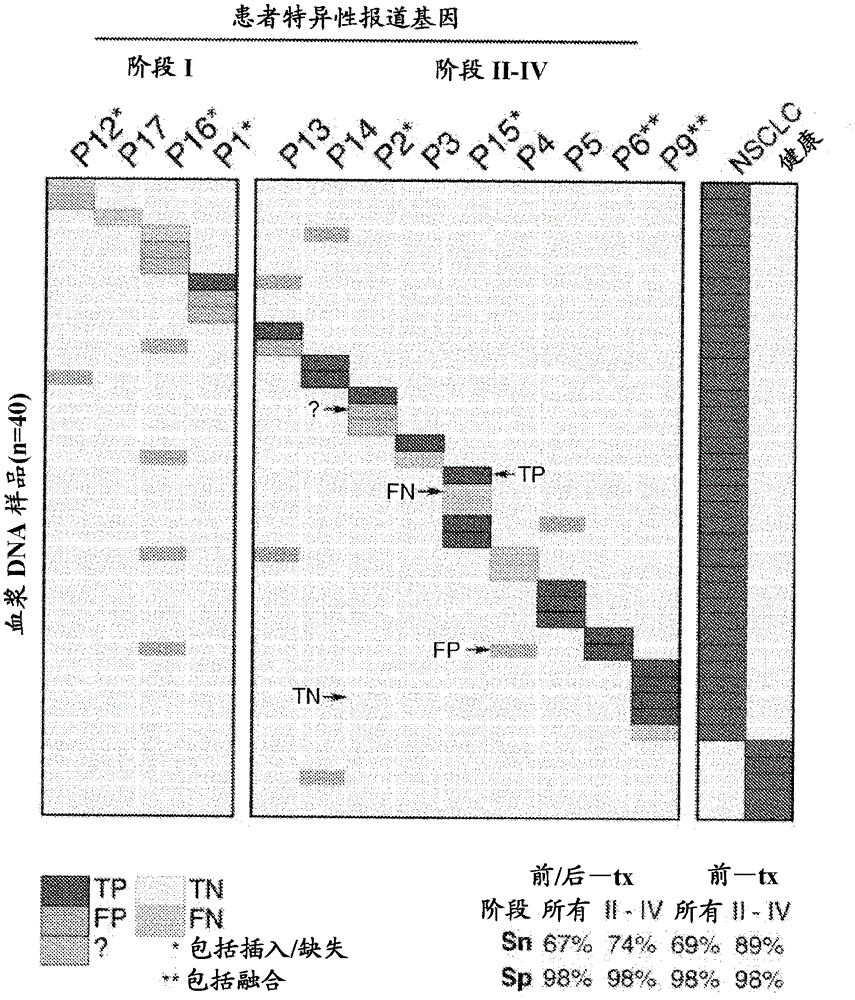
[0783] Circulating tumor DNA (ctDNA) represents a promising biomarker for noninvasive detection of disease burden and recurrence monitoring. However, existing ctDNA detection methods are limited in sensitivity, focus on a small number of mutations, and / or require patient-specific optimization. In order to solve these shortcomings, the Deep Sequencing Cancer Personalized Profiling Analysis (CAPP-Seq) has been developed, an economical and highly sensitive method for quantifying plasma ctDNA in almost every patient. We performed CAPP-Seq on non-small cell lung cancer (NSCLC), designed to identify> 95% of tumors are mutated, and point mutations, insertions / deletions, copy number variants and rearrangements are also detected. When the tumor mutation characteristics are known, we detected ctDNA in 100% stage II-IV NSCLC pretreated plasma samples and 50% stage I NSCLC samples, a...
Embodiment 2
[0881] Example 2: Design an individualized selector set
[0882] In some cases, it may be impractical to monitor the tumor burden in patients with known cancers using "existing" strategies. This strategy uses knowledge from a group of patients of the same tumor type and uses CAPP-Seq to select Sexually capture genomic regions frequently mutated in this tumor type. These conditions include, but are not limited to, the following cases, where (1) the tumor has an unknown original histology (for example, CUP); (2) the histology is known, but is too rare to have enough of the previously described tumor types The number of patients defines the average patient tumor somatic gene panorama (for example, subtype of soft tissue sarcoma); (3) the histology is known, but the average / median recurrence somatic damage in this tumor type is too large It is too low to reach the expected sensitivity level (for example, pediatric tumors, etc.); or (4) The histology is known, and the average / median ...
Embodiment 3
[0884] Example 3. Use of selector set in the diagnosis of cancer
[0885] Ingredients
Volume (μL)
cfDNA
1-75
Phosphorylation reaction buffer (10X)
10
5
5
dNTPs
4
DNA polymerase I, large (Klenow)
1
139 --> Sterile H 2 O
-Add to a total volume of 100μL
[0886] The end repair reaction mixture was incubated in a thermal cycler for 30 minutes at 20°C.
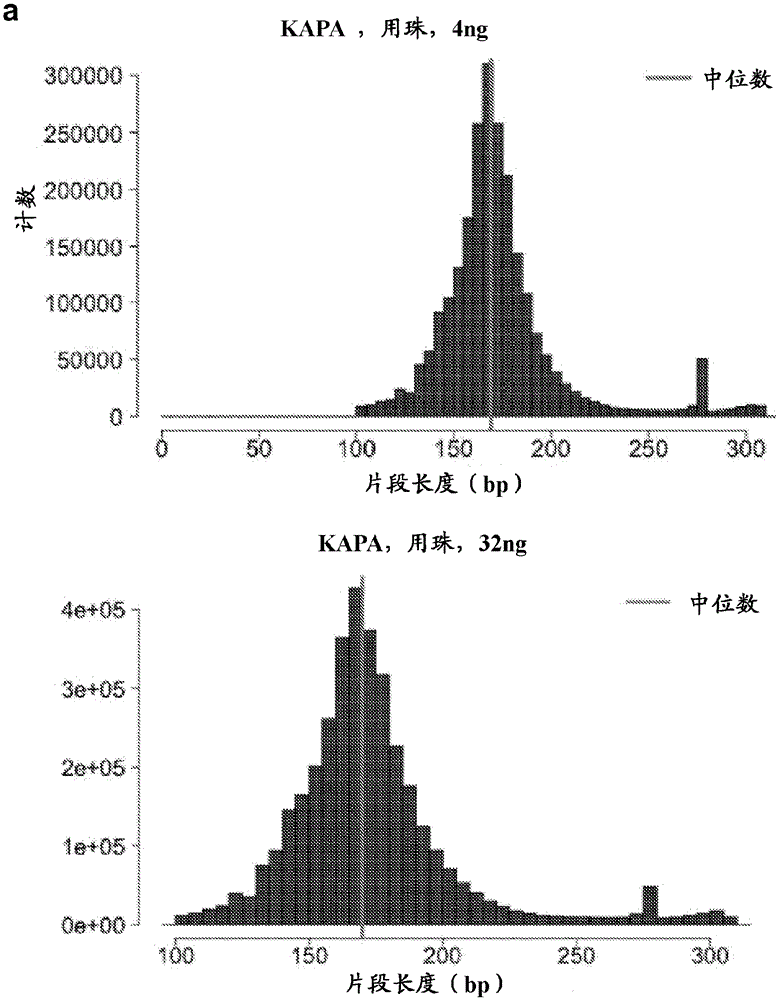
[0887] The purification of the end-repaired cfDNA was performed by adding 160 μL (1.6X) of resuspended AMPureXP beads to the end-repair reaction mixture. Mix the AMPure beads on a vortex mixer or pipette up and down (for example, 10 times or more) into the solution. The reaction was incubated at room temperature for 5 minutes. The reaction was placed on a magnetic stand to separate the beads from the supernatant. After the solution is clear (about 5 minutes), the supernatant is removed and discarded. The beads were washed twice by adding 200 μL of 80% freshly prepared e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com