Nest type PCR detection method for different variants of ostreid herpes virus
An oyster herpes virus, the technology to be detected, applied in the direction of microbial determination/inspection, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem of inability to accurately detect the existence of mutant strain infection and the inability of OsHV-1 detection primers Combination and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
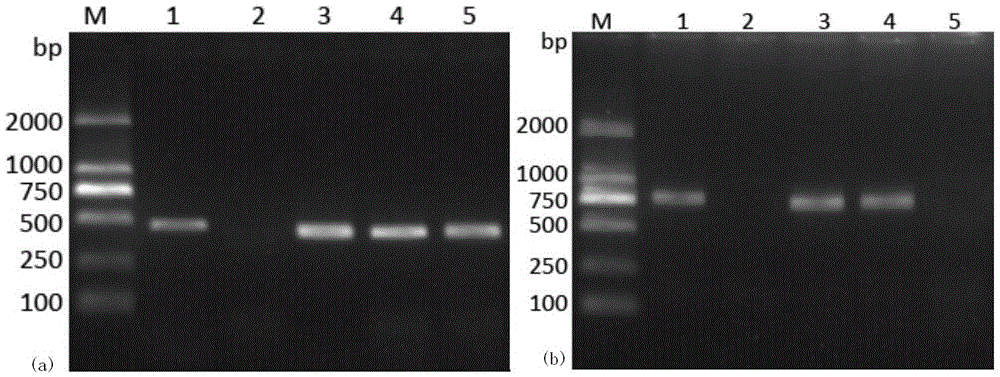
[0028] Embodiment 1: PCR primer design
[0029] Using the Primer5.0 primer design software, based on the nucleotide sequence of the key functional domain of the DNA polymerase gene, the nested PCR detection primers were designed: the upstream sequence of the outer primer, Pol-F: 5'-GATTTCAACTCGCAATACC-3', the outer primer The downstream sequence of the primer, Pol-R: 5'-GGCAGACACAGATTCCACA-3', the amplified fragment size is 573bp; the upstream sequence of the internal primer, nPol-F: 5'-GTGTTTCTACATTGCTGG-3', the downstream sequence of the internal primer nPol-R: 5' -CTGTTGGCGTTGACCTTC-3', the size of the amplified fragment is 386bp.
[0030] Optimization of the first-round PCR reaction system and amplification program: 2 μL of DNA from a scallop sample known to be infected with OsHV-1 was used as a positive template, DNA polymerase (5U / μL) was 0.4 μL, 10×PCR buffer was 5.0 μL, dNTPs (2.5 mmol / L) 4.0μL, Mg 2+(25.0mmol / L) 4.0μL, upstream and downstream primers (10.0μmol / L) 2....
Embodiment
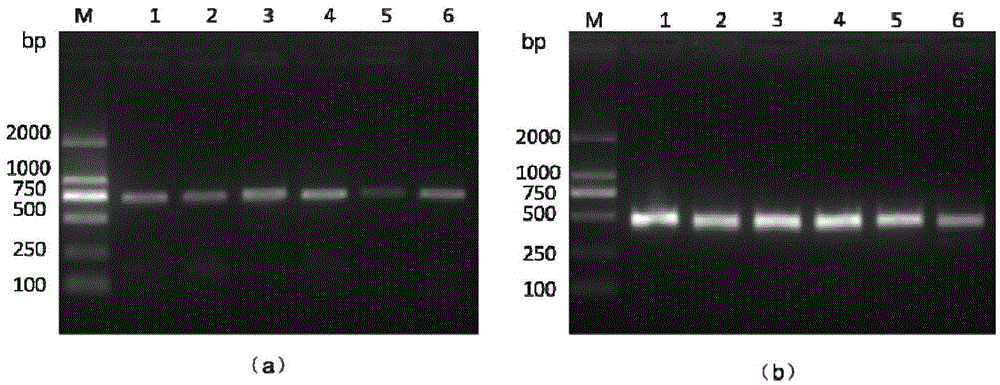
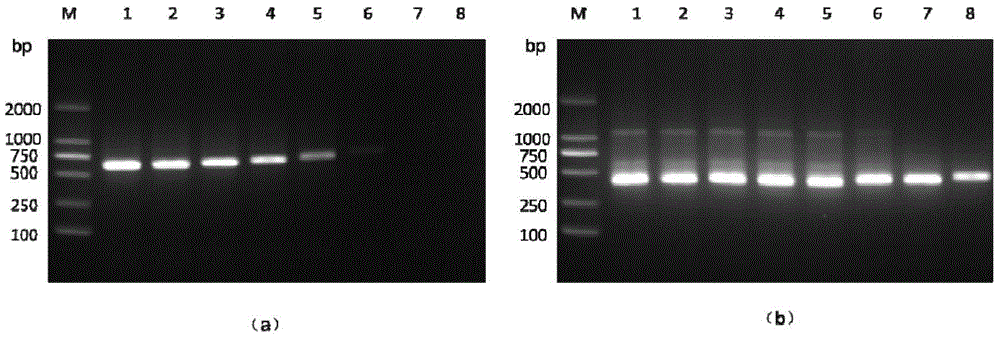
[0036] Embodiment: Nested PCR detection method detects different shellfish OsHV-1 infection situation
[0037] (1) DNA extraction: Take about 30 mg of the mantle tissue of different species of shellfish disease materials collected in different years and collected in the laboratory, and use the marine animal tissue genome kit of Tiangen Biochemical Technology (Beijing) Co., Ltd., provided according to the kit instructions The steps are to extract sample DNA and use it as a template for nested PCR detection. The nucleic acid of Chlamys farreri positive for OsHV-1 was used as a positive control, and the nucleic acid of Chlamys scallop negative for OsHV-1 was used as a negative control.
[0038] (2) The first round of PCR reaction: using TaKaRaEx of Treasure Bioengineering (Dalian) Co., Ltd. Add the following reagents to a 50 μL PCR reaction system: DNA polymerase (5U / μL) 0.4 μL, 10×PCR buffer 5.0 μL, dNTPs (2.5 mmol / L) 4.0 μL, Mg 2+ (25.0mmol / L) 4.0μL, upstream and downstream ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com