Method for preparing RNA (ribonucleic acid) probe by using miR-155 precursor as template
A technology of RNA probe and RNA polymerase, which is applied in the field of preparing RNA probes to achieve the effect of reducing background signal, reducing background signal interference, and shortening experiment time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
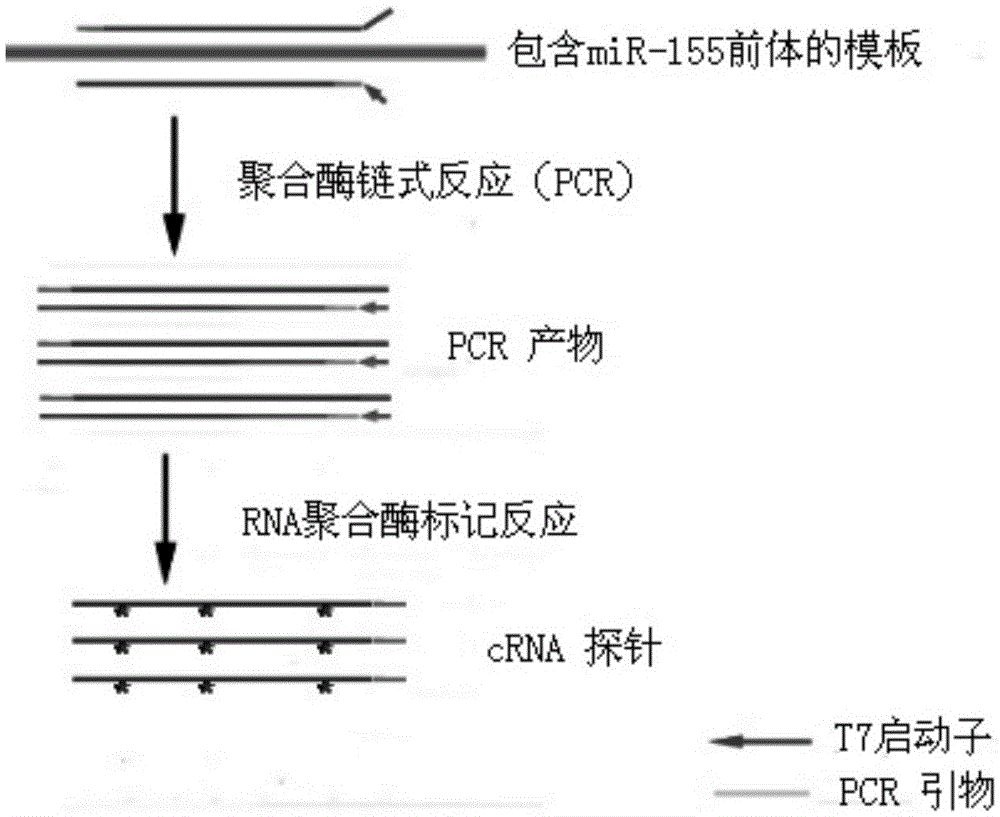
[0042] Example 1: Preparation of RNA probes using miR-155 precursor as a template
[0043] The process of preparing RNA probes using miR-155 precursor as a template can be found in figure 1 .
[0044] 1. Design of PCR primers:
[0045] (1) Find the name and sequence of its corresponding pre-miRNA in the miRBase database (http: / / www.mirbase.org / ), namely the miR-155 precursor (AccessionMI0000681): HomosapiensmiR-155stem-loop.
[0046] (2) Query the sequence of the corresponding pre-miRNA, and the result shows that the length of the pre-miRNA is about 100nt.
[0047] (3) Design a pair of primers.
[0048] Primer design:
[0049]
[0050] The primers mentioned in the above table are only examples, and those skilled in the art can design more suitable primers according to the principle of the present invention.
[0051] (4) Add the T7 promoter sequence to the 5' end of the reverse primer (see the table above) to ensure that the PCR product is complementary to the target fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com