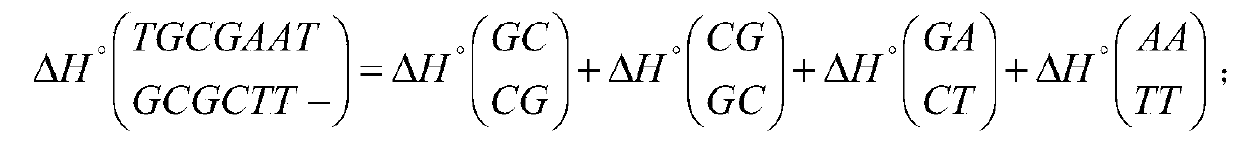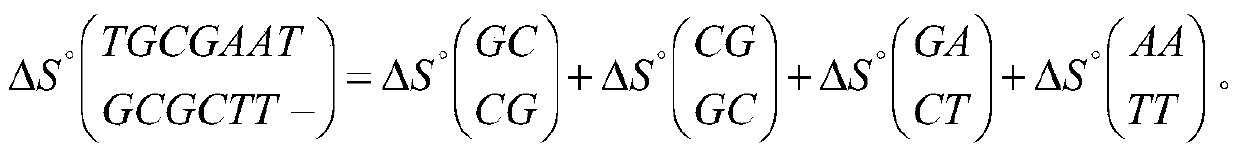Method and system for deducing bonding site of oligonucleotide on genome
A technology of oligonucleotides and binding sites, applied in the field of inferring oligonucleotide binding sites, which can solve problems such as low efficiency, complex calculations, and slow search processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
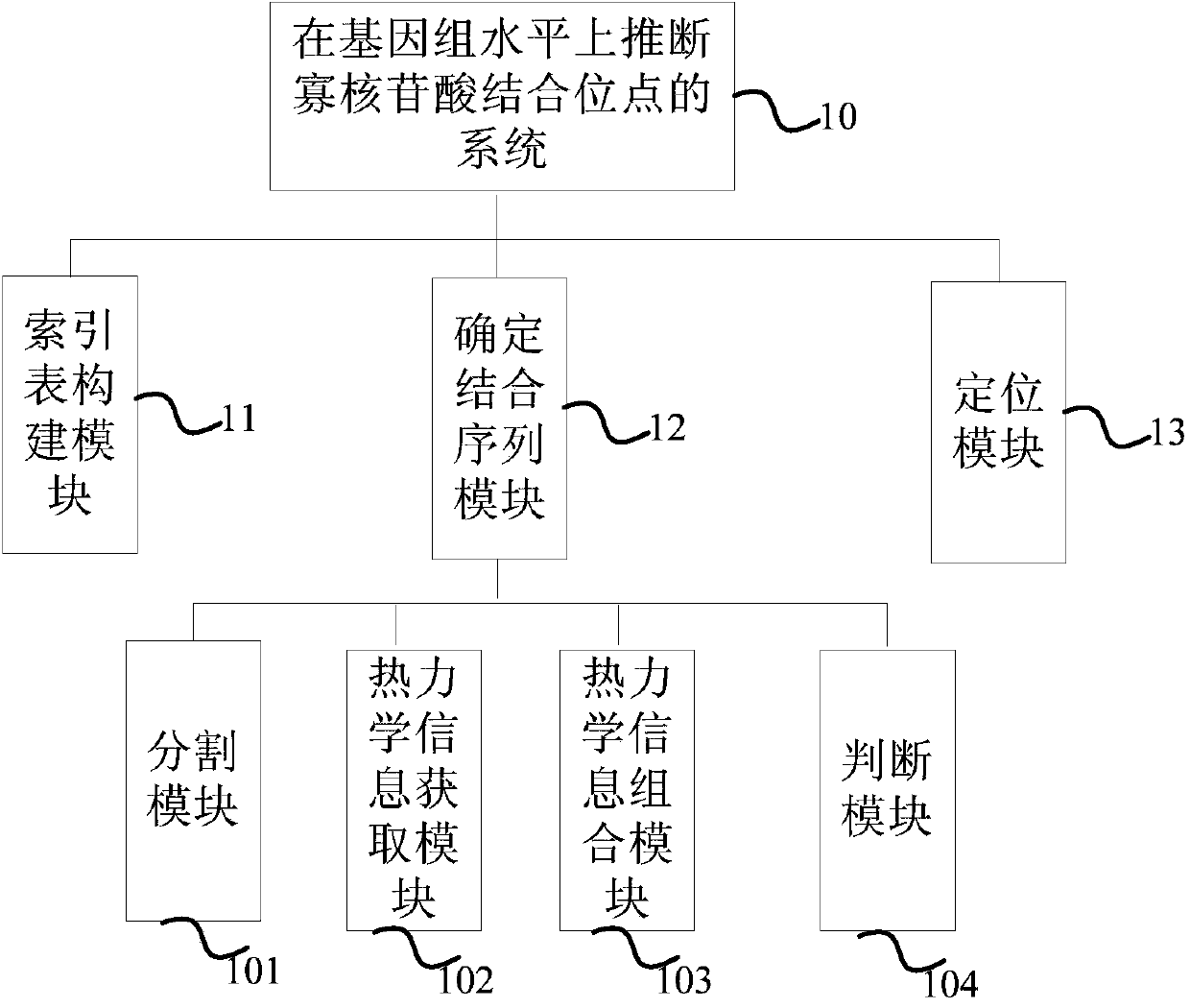
[0088] Example 1: Using figure 1 The determinative binding sequence module 12 in the system 10 for estimating oligonucleotide binding sites at the genome level as shown obtains the binding sequence of the oligonucleotide sequence to be deduced according to the method of the present invention
[0089] Step 1: use the index table construction module 11 in the system 10 to construct an index table of the thermodynamic information of any 7-mer oligonucleotide, the thermodynamic information is the pairwise hybridization of the oligonucleotide and all its binding sequences Information, including hybrid structure, hybrid sequence, enthalpy, entropy, and free energy;
[0090] 1) Each base (such as A) on any oligonucleotide will encounter five situations (T, A, G, C, -) in the binding sequence, so that 0, 1 in the pentadic number , 2, and 3 respectively correspond to one of the four kinds of deoxyribonucleic acid, and 4 corresponds to the gap. Any 7-mer oligonucleotide and all its bin...
Embodiment 2
[0131] Example 2 Use the positioning module 13 in the application system 10 to locate the position of the thermodynamically stable binding sequence obtained in Example 1 on the whole human genome
[0132] The known k-mer algorithm processing process is as follows:
[0133] (1) Using the sliding window method, set the window size to k-mer, and determine a total of 4 segments to be searched k sequence, where k=9,
[0134] (2) With a step size of 1, scan the 9-mer fragment to be searched from front to back on the specified genome,
[0135] (3) Record the positions of the 9-mer fragments to be searched on the positive and negative strands of different genes,
[0136] (4) Repeat (2) (3) process for all segments to be searched,
[0137] (5) Express the arbitrary 9-mer fragments appearing in the genome in the form of decimal numbers, and store the position information of each arbitrary 9-mer fragment on the genome together in the database.
[0138] The k-mer preprocessing algorit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com