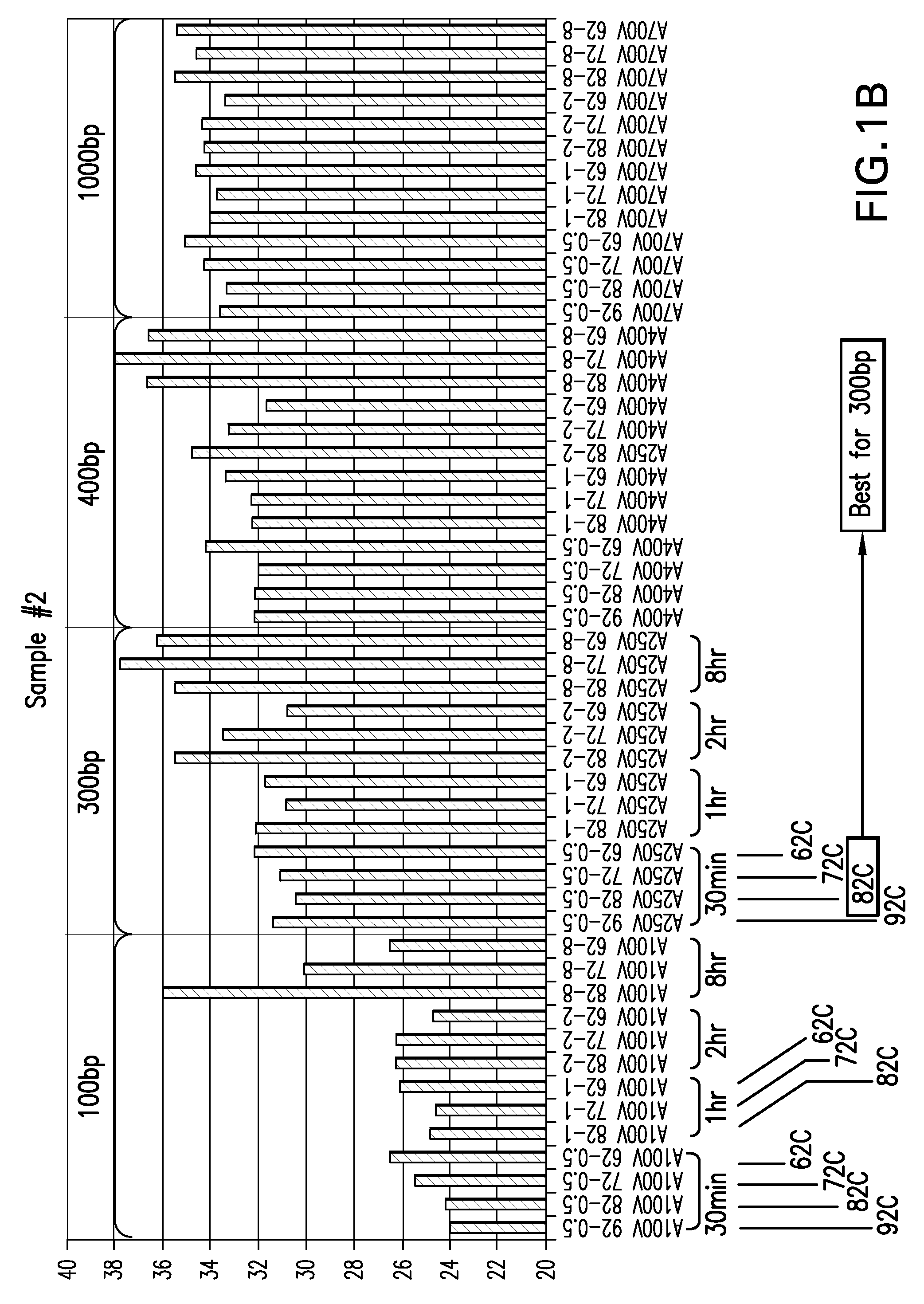
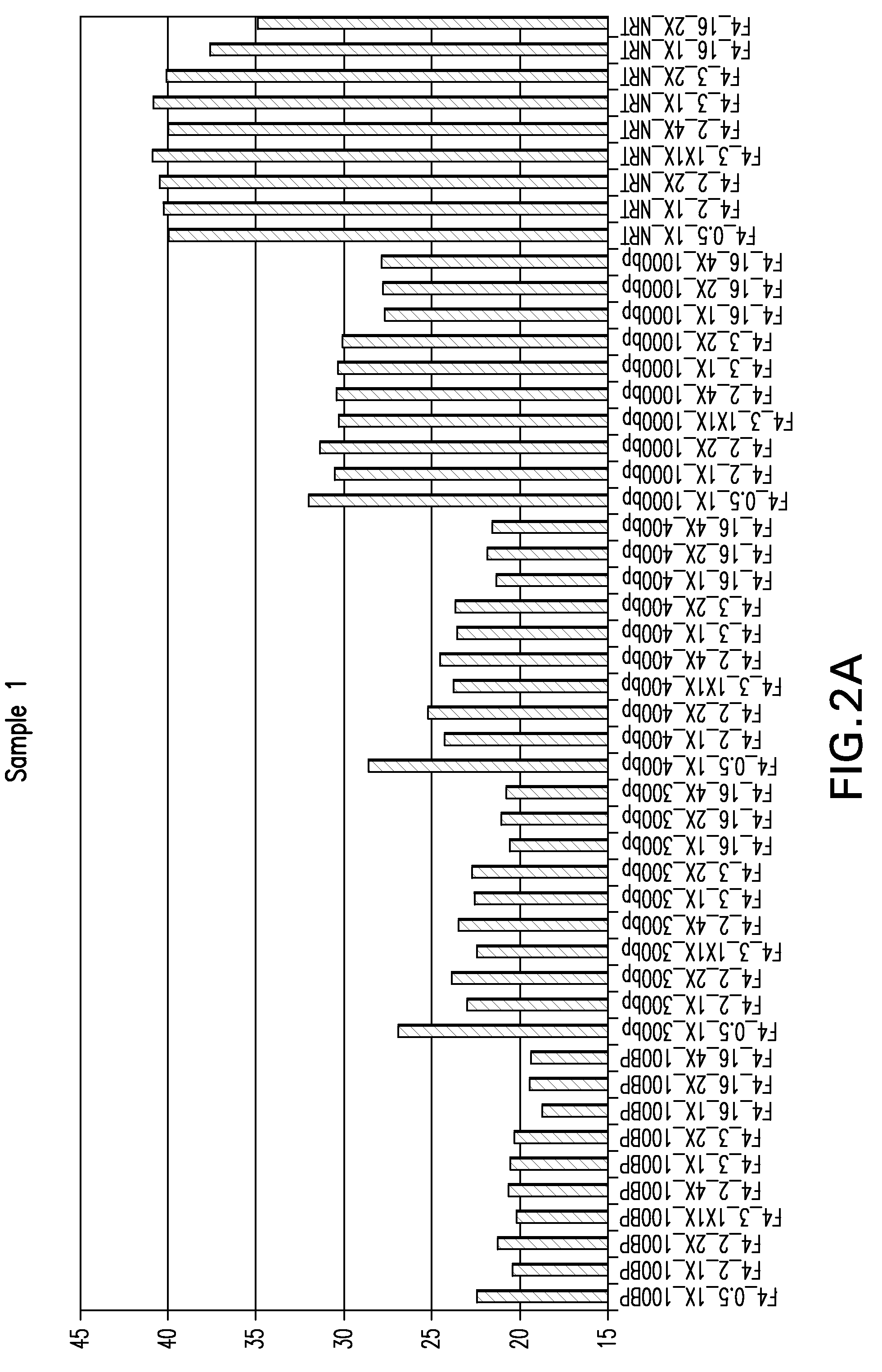
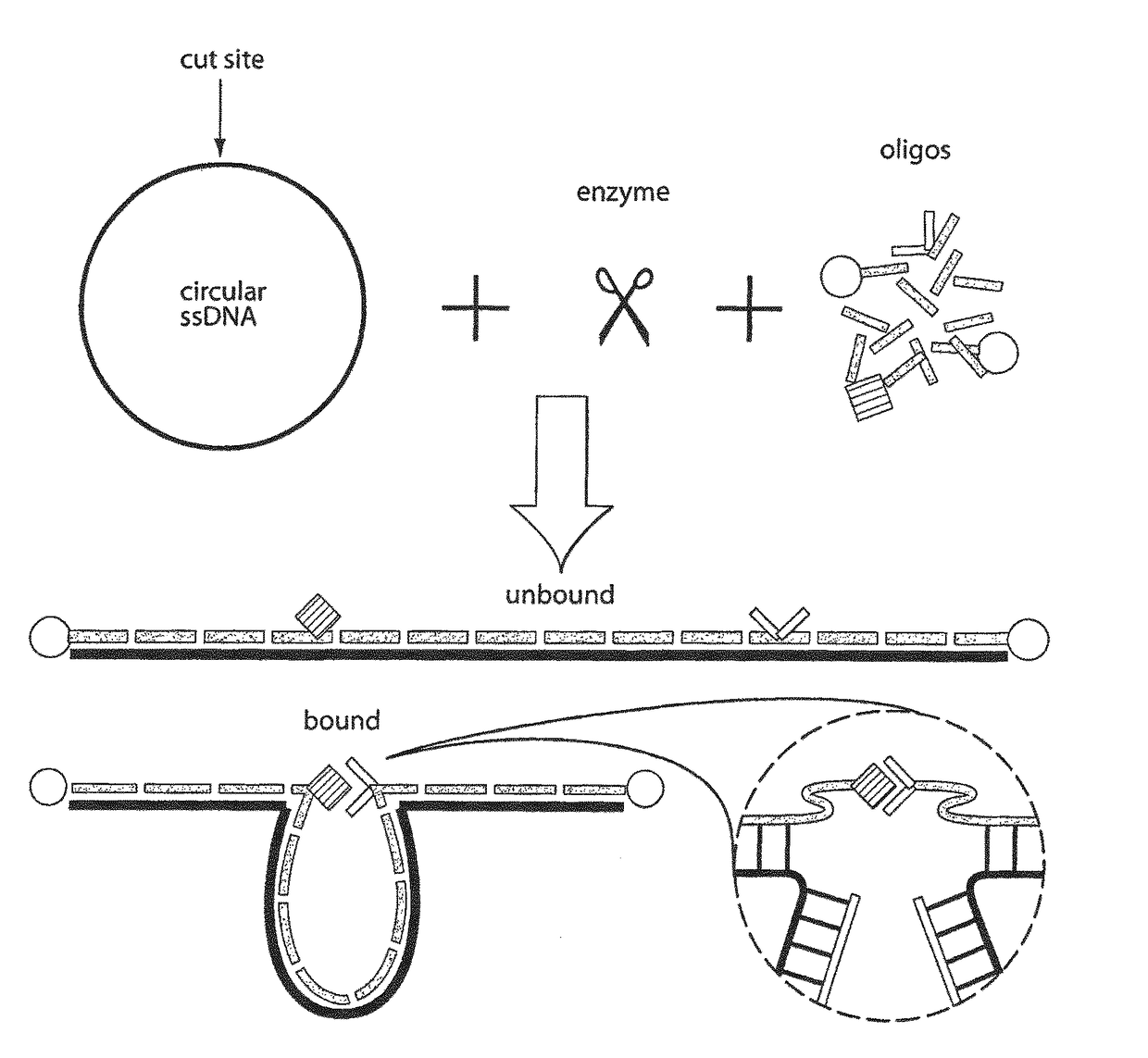
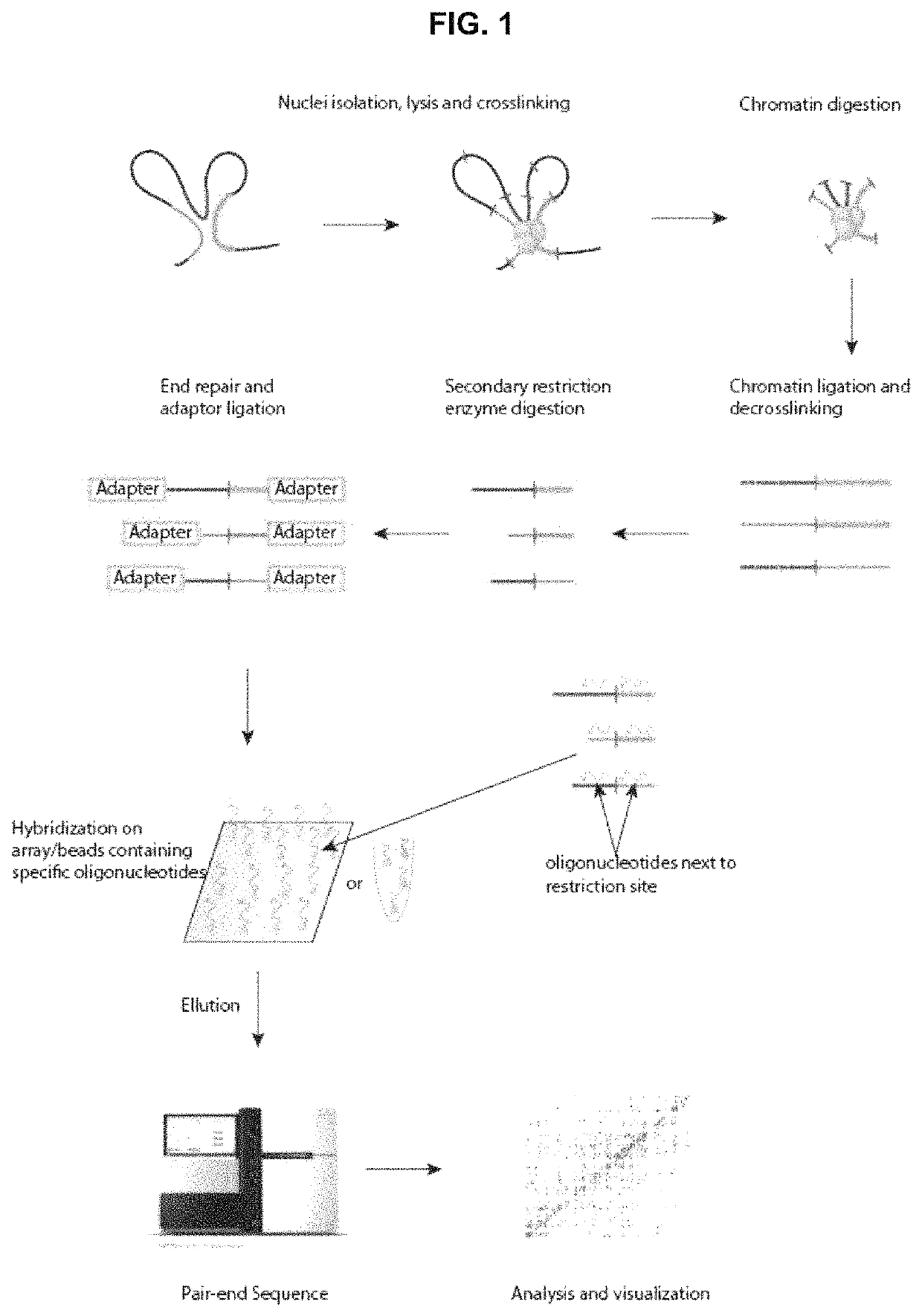
Patents
Literature
84 results about "Oligonucleotide hybridization" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
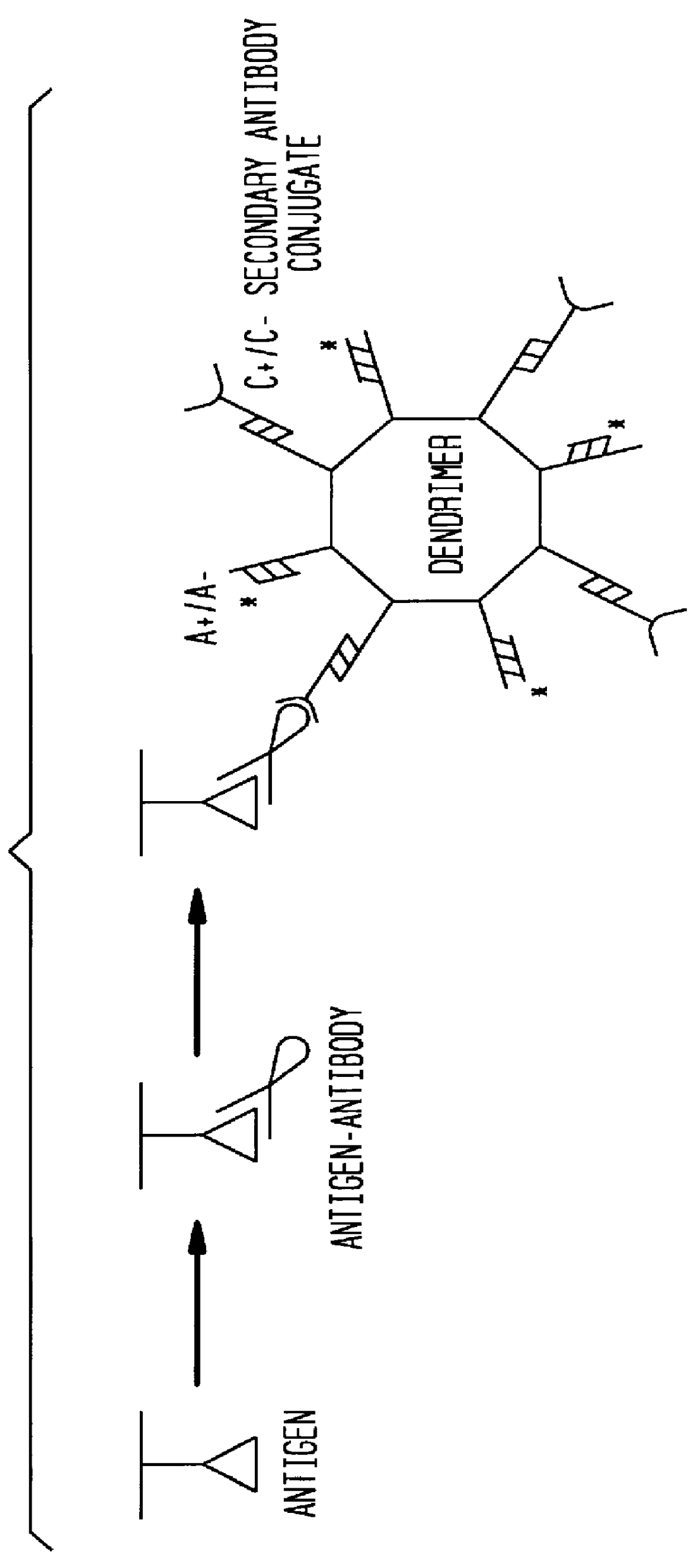
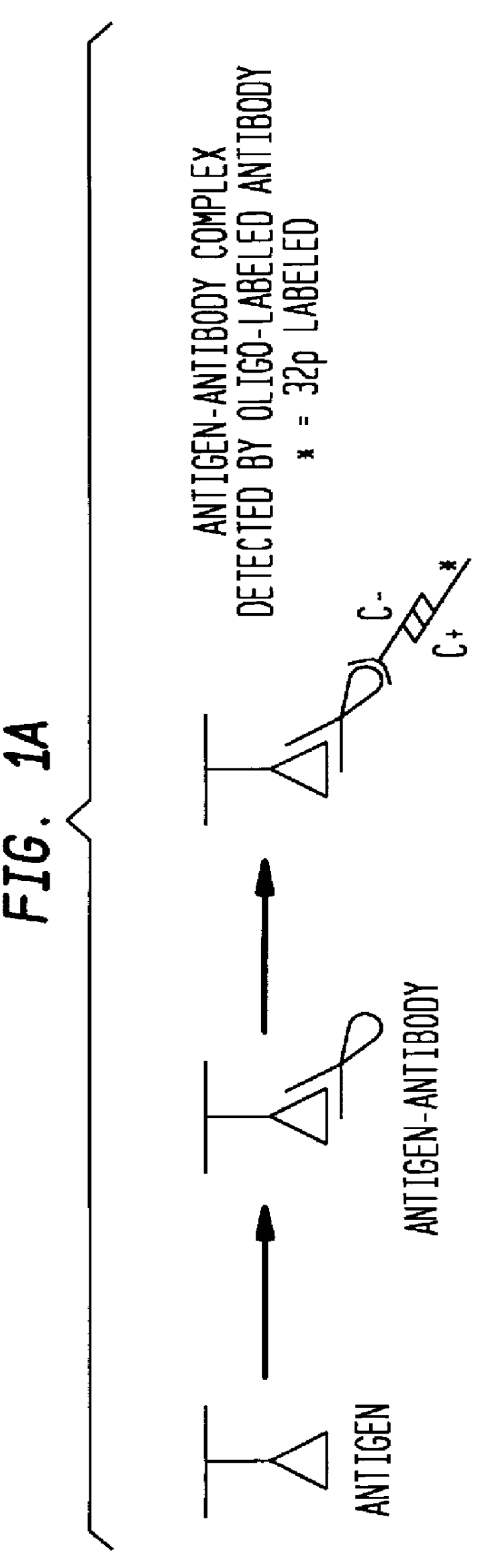
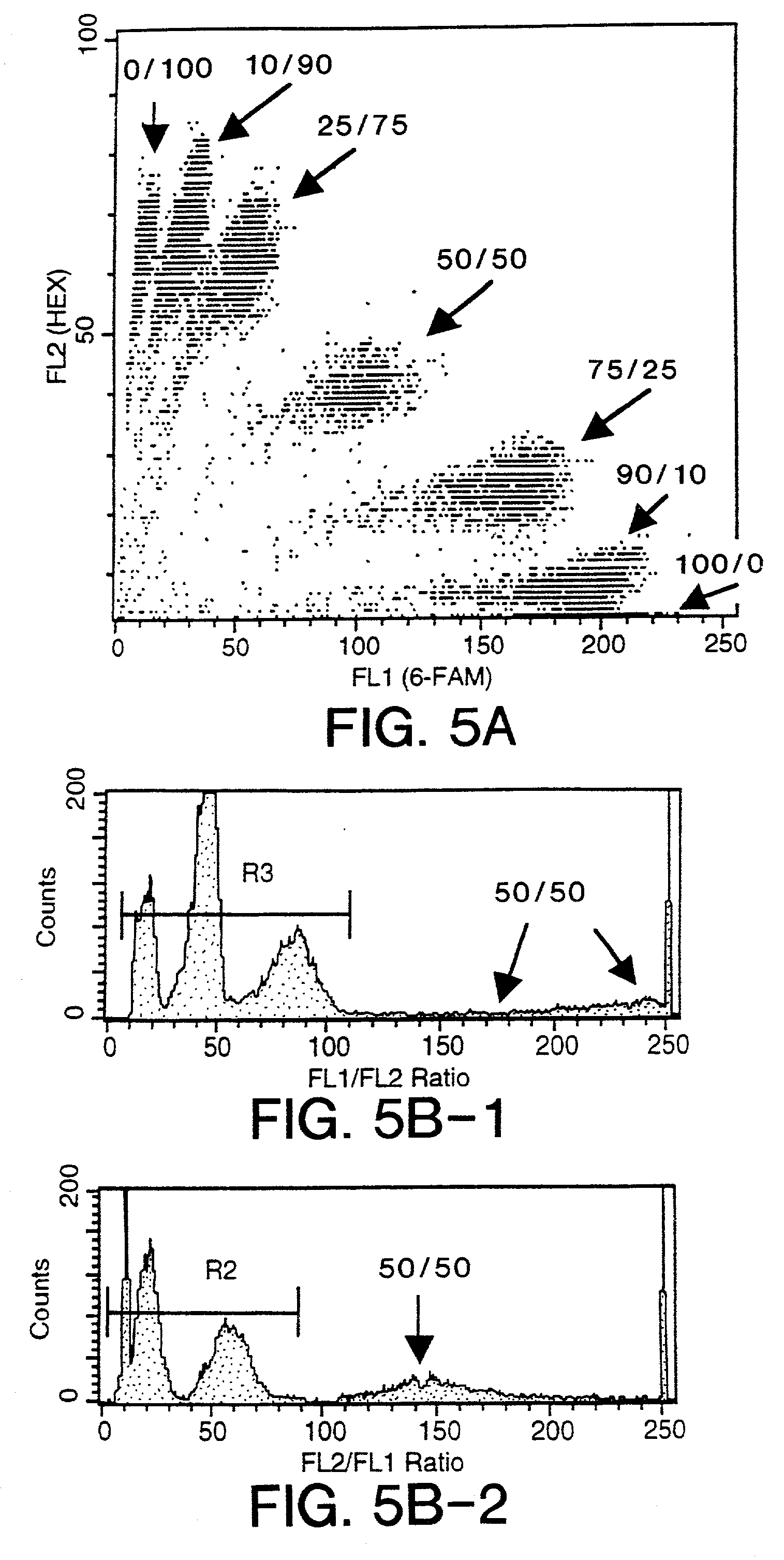
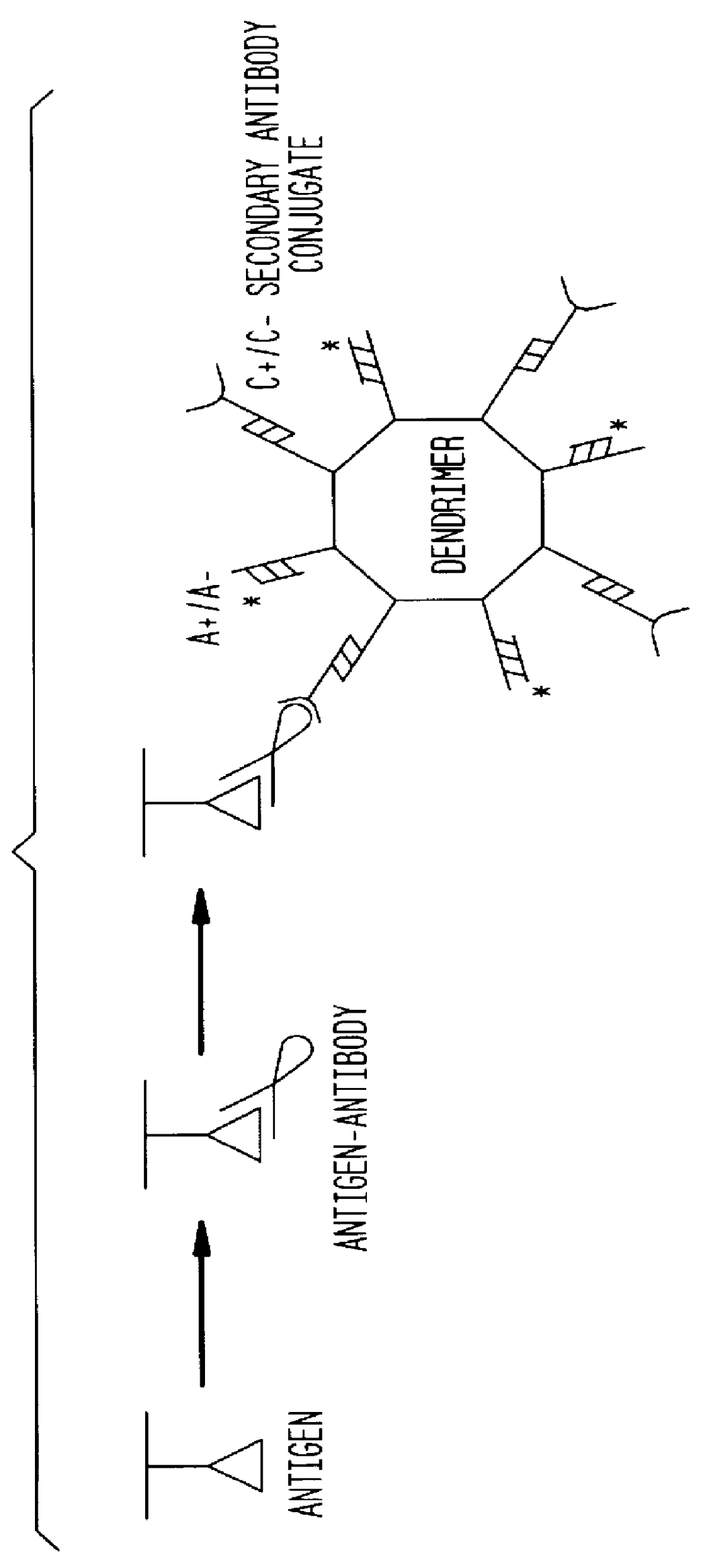
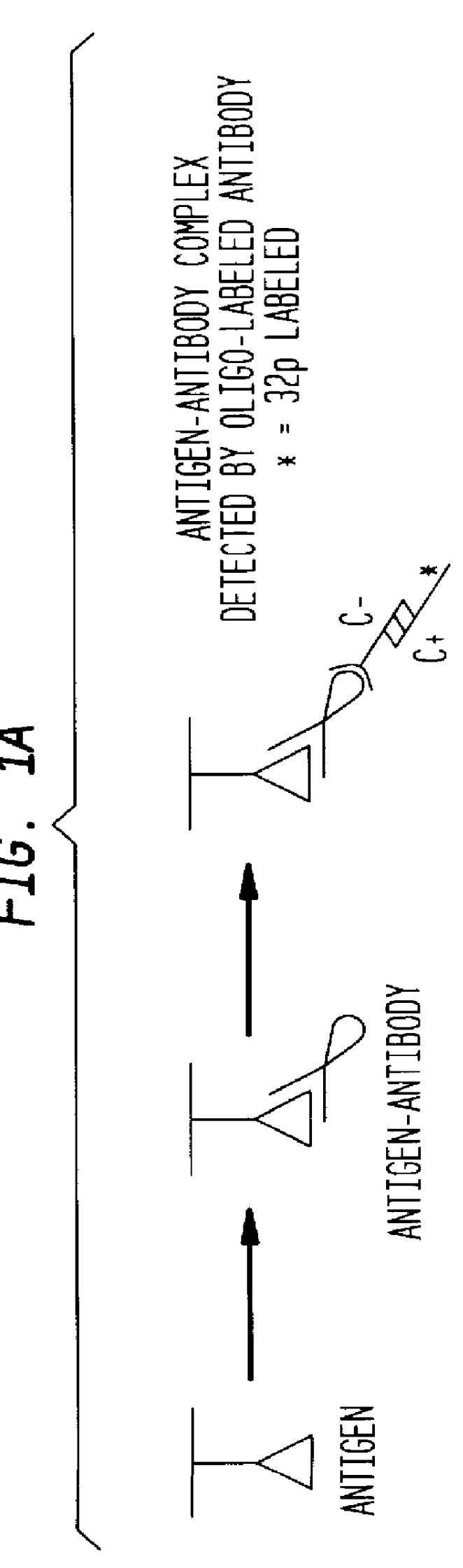
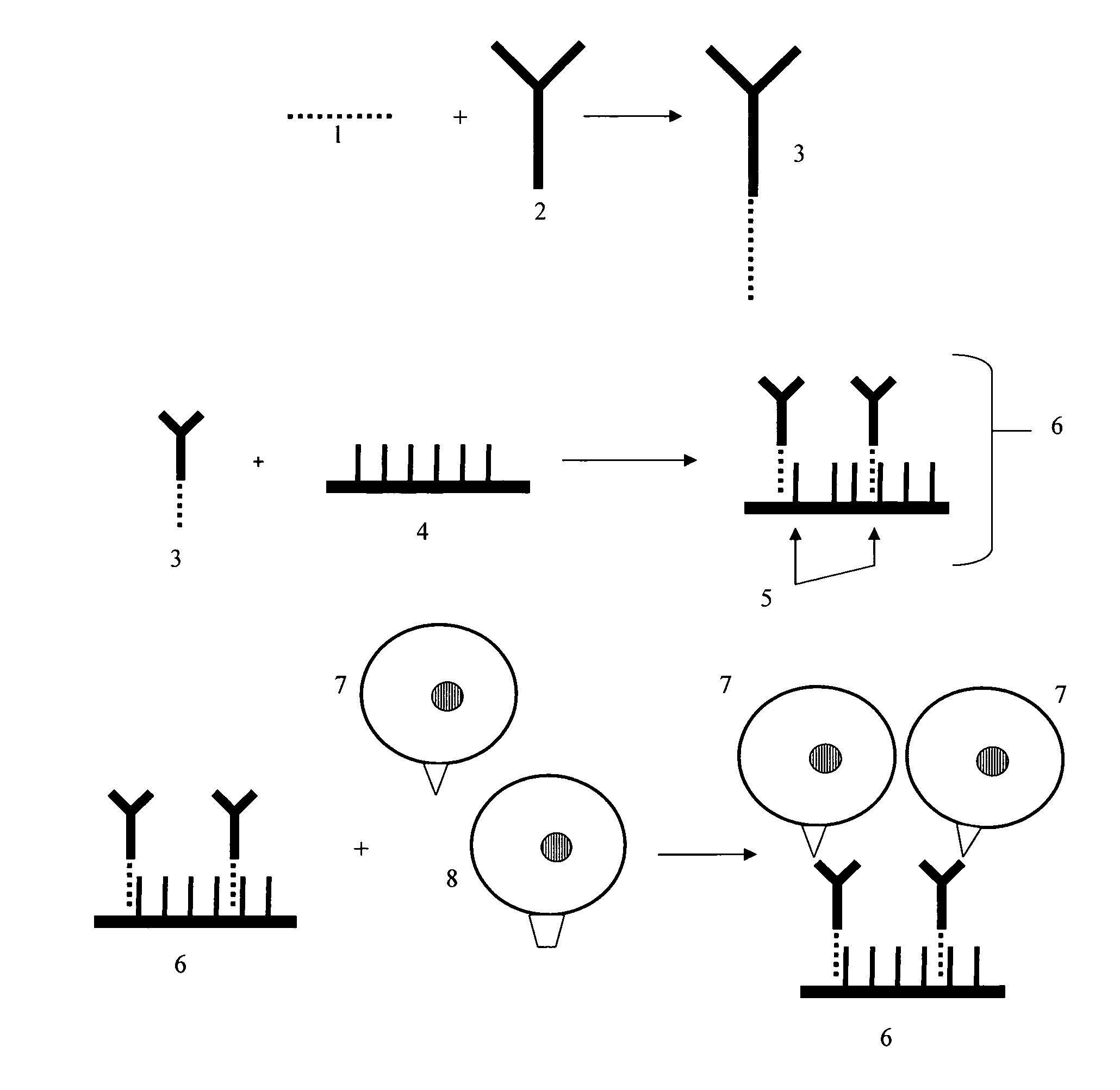
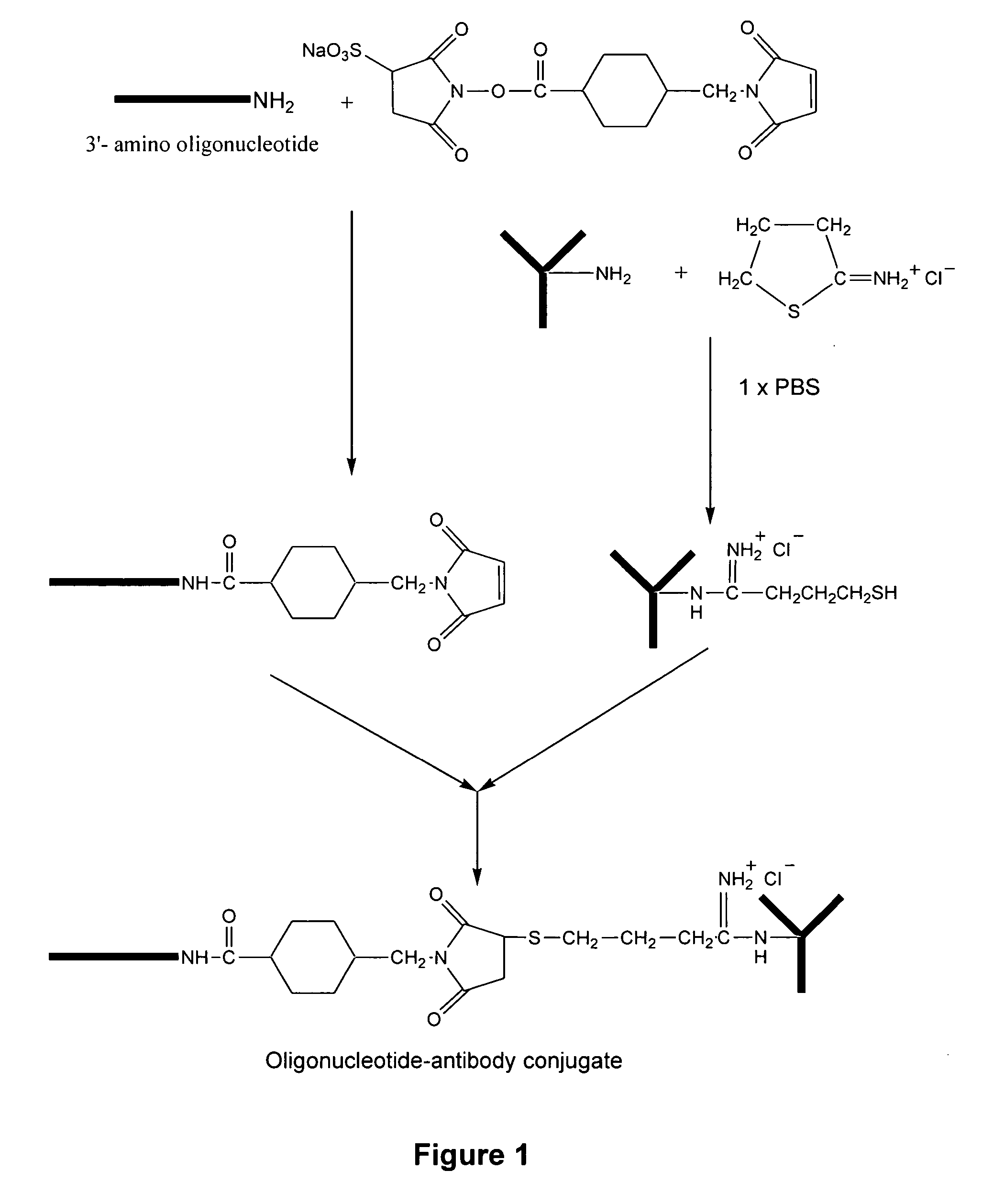
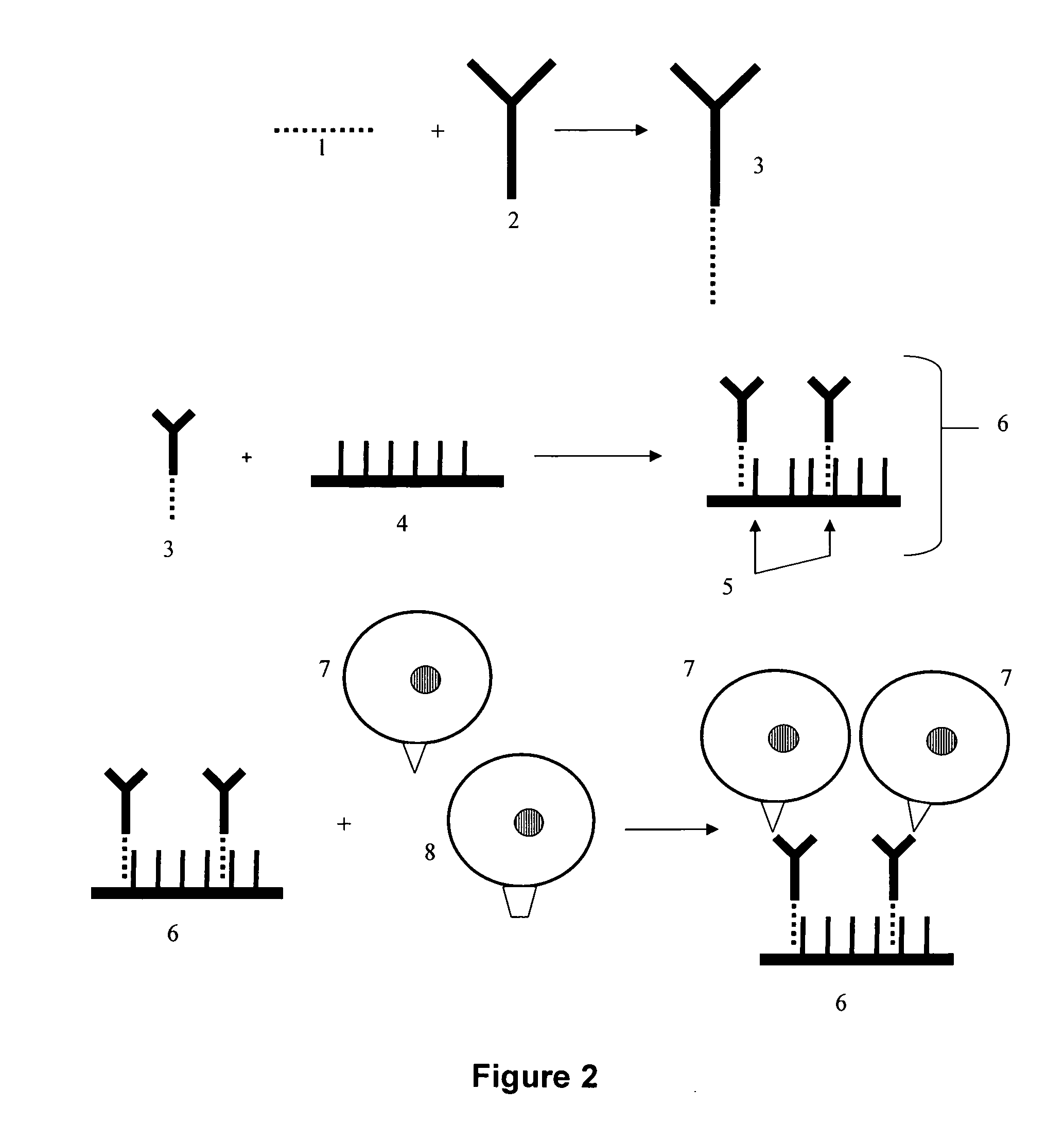
Detection of antigens via oligonucleotide antibody conjugates
InactiveUS6117631AHigh detection sensitivityEnhancing observed signalIn-vivo radioactive preparationsSugar derivativesDendrimerAntibody conjugate
The present invention provides a method of detecting antigens, which comprises immobilizing an antigen to a solid support and contacting the solid support with a means for hybridizing a labeled dendrimer to the antibody, through an oligonucleotide complexed thereto. A directly oligonucleotide labeled primary antibody or an oligonucleotide labeled secondary antibody may be employed, and a conventionally labeled dendrimer can subsequently be hybridized to the oligonucleotide through one or more of the outer arms of the dendrimer. The present invention offers the advantage over conventional methods of antigen detection by providing multiple label molecules per antigen, thereby enhancing the observed signal associated with the label.
Owner:GENISPHERE LLC
Methods for measuring relative amounts of nucleic acids in a complex mixture and retrieval of specific sequences therefrom
InactiveUS20020172965A1Poor enrichmentMultiplicative improvement in overall enrichmentMicrobiological testing/measurementOther foreign material introduction processesCDNA libraryNucleic acid sequencing
The present invention relates to a method for the comparative assessment of the level of specific nucleic acid sequences in samples derived from different sources. More specifically, the invention relates to a method using oligonucleotides covalently linked to a solid support, such as beads, to isolate specific labeled nucleic acid sequences from complex mixtures. The methods disclosed allow quantitative comparisons of the amount of nucleic acid of defined sequence in a plurality of different samples of nucleic acid, e.g., from different cells or tissues or from genetic libraries. Nucleic acids from the samples are labeled in such a fashion that the signals can be distinguished and compared following hybridization to the oligonucleotides on the beads. According to the invention, the solid supports with the hybridized nucleic acid may be retrieved, and the target nucleic acid eluted and analyzed. Furthermore, the invention provides a method for tagging individual clones from a cDNA library such that they can be identified uniquely and retrieved by hybridization to specific beads.
Owner:DELTAGEN PROTEOMICS
Detection of antigens via oligonucleotide antibody conjugates
InactiveUS6110687AHigh detection sensitivityEnhancing observed signalSugar derivativesMicrobiological testing/measurementDendrimerAntibody conjugate
The present invention provides a method of detecting antigens, which comprises immobilizing an antigen to a solid support and contacting the solid support with a means for hybridizing a labeled dendrimer to the antibody, through an oligonucleotide complexed thereto. A directly oligonucleotide labeled primary antibody or an oligonucleotide labeled secondary antibody may be employed, and a conventionally labeled dendrimer can subsequently be hybridized to the oligonucleotide through one or more of the outer arms of the dendrimer. The present invention offers the advantage over conventional methods of antigen detection by providing multiple label molecules per antigen, thereby enhancing the observed signal associated with the label.
Owner:GENISPHERE LLC
Real-time PCR microarray based on evanescent wave biosensor
InactiveUS20060088844A1Bioreactor/fermenter combinationsBiological substance pretreatmentsWavelengthNatural abundance
A system and method for simultaneous, quantitative measurement of nucleic acids in a sample. Fluorescently tagged amplicons of the target nucleic acids are localized on a substrate surface by hybridization to oligopobes that have been arrayed and tethered to the substrate surface in a pre-determined, two-dimensional pattern. The hybridized, amplicons are then detected by exciting their fluorescent tags using an evanescent wave of light of the appropriate wave-length. Because of the limited penetration of the evanescent wave (about 100-300 nm), the fluorescently tagged nucleotides in the remainder of the reaction cell do not fluoresce. By measuring the fluorescence at various locations on the substrate surface, the current abundance of hybridized amplicons of each of the target nucleic acids can be determined. The analytic techniques of real time PCR may then be used to obtain accurate, quantitative measurements for each of the nucleic acids in the sample.
Owner:HONEYWELL INT INC
Nucleic acid detection method
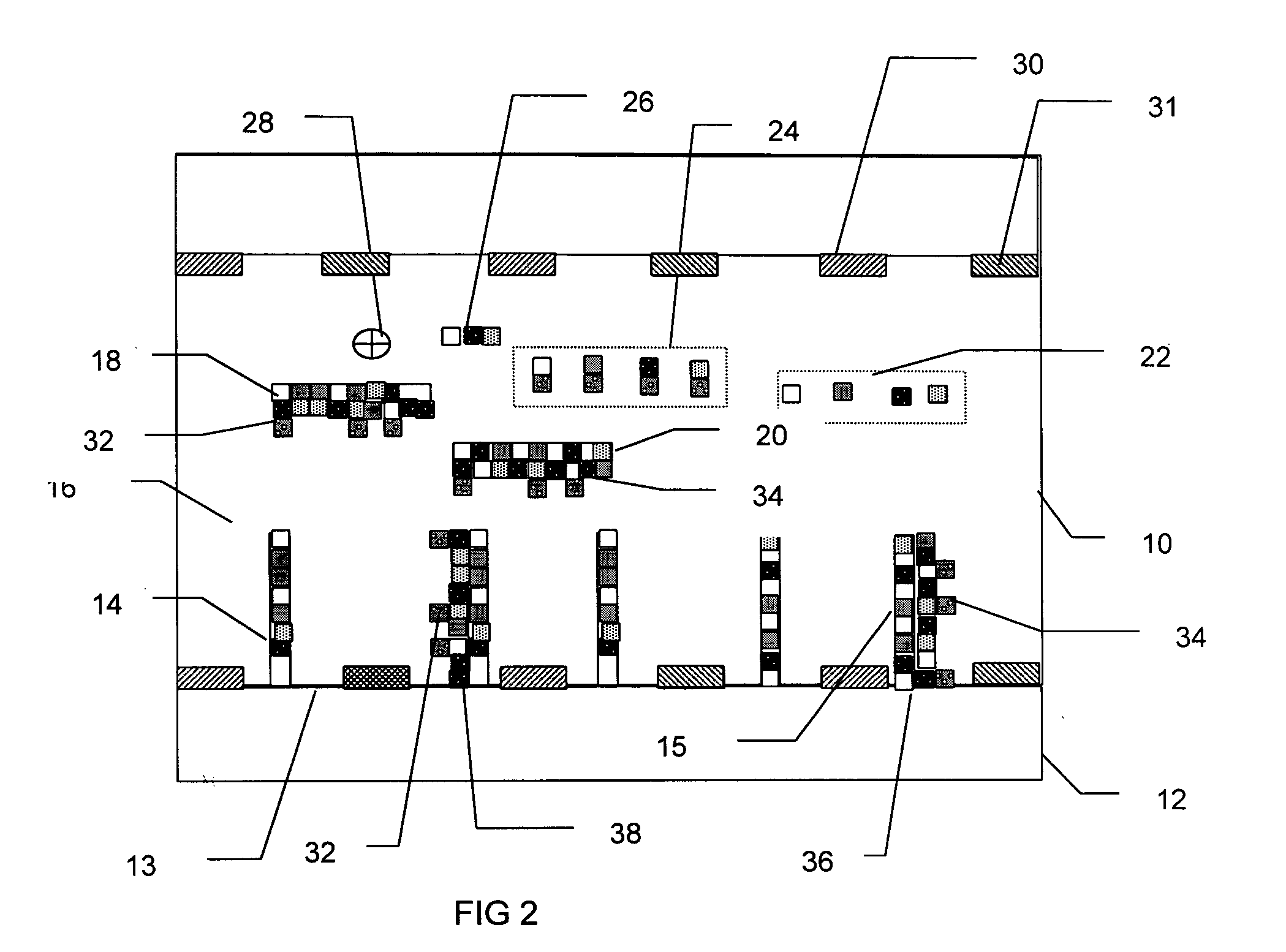
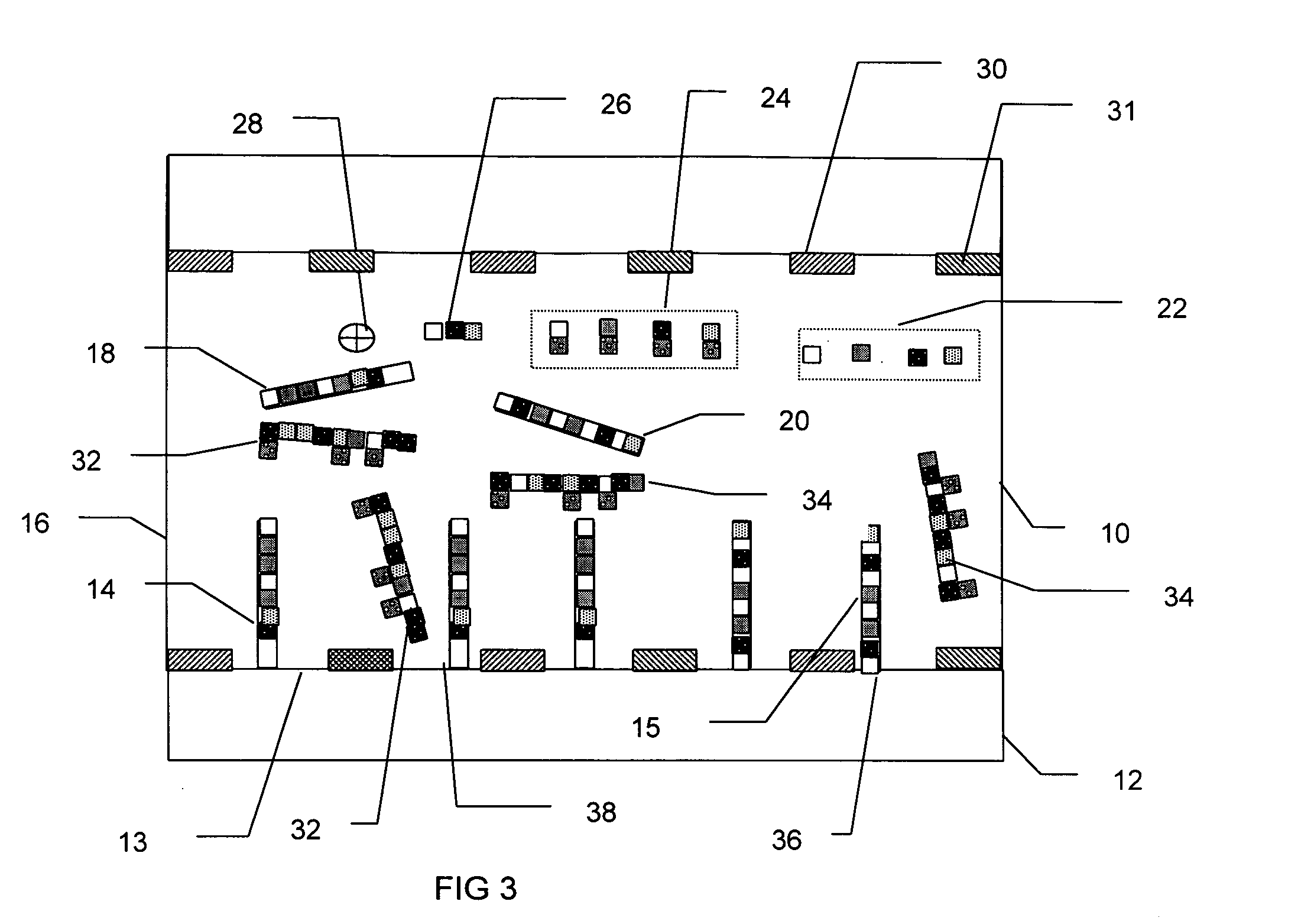
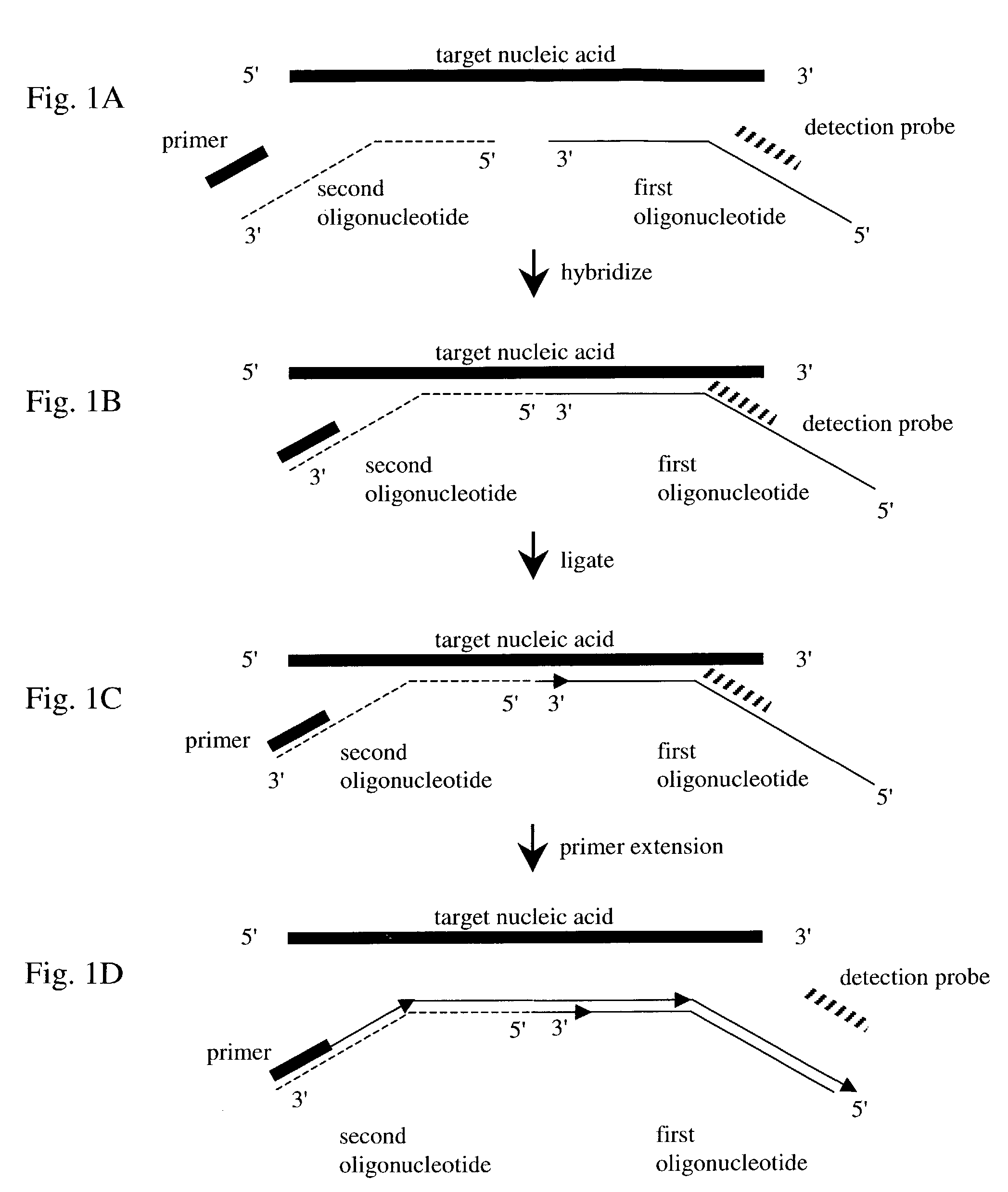
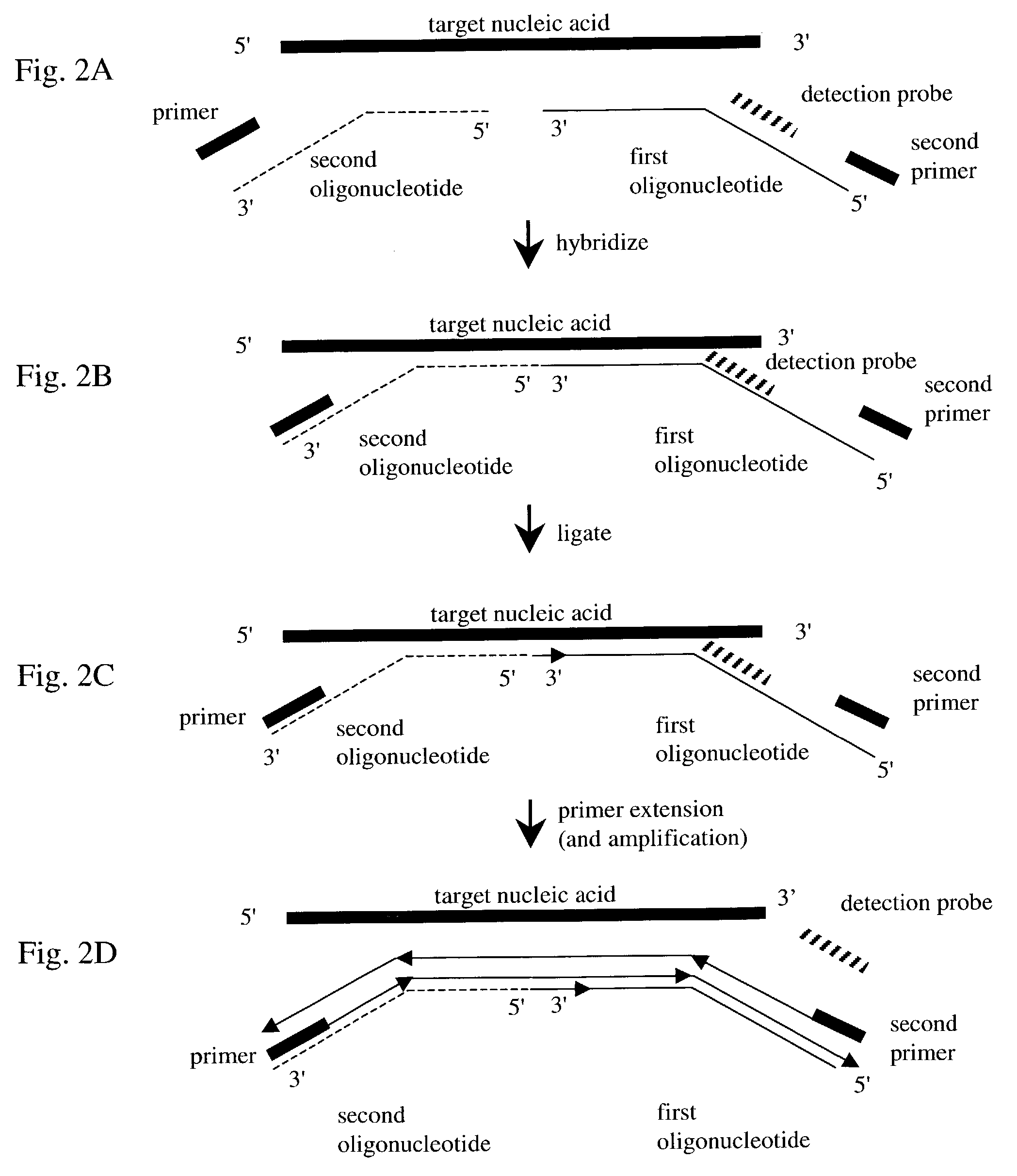
InactiveUS7282355B2Rapid and reliable and cost-effectiveDetect presenceSugar derivativesMicrobiological testing/measurementNucleic acid detectionNucleotide sequencing
Disclosed are methods for detecting a target nucleic acid molecule having a known sequence among a plurality of nucleic acid molecules. The method includes hybridizing two oligonucleotides to adjacent nucleotide sequences on the target nucleic acid molecule, ligating the two oligonucleotides together, and extending a primer that hybrizes to the 3′ end of the oligonucleotide that hybrized to the 5′ nucleotide sequence on the target nucleic acid molecule. If the target nucleic acid molecule is present (and thereby allowing ligation of the two oligonucleotides), extension of the primer dislodges a detection probe hybridized to the 5′ end of the oligonucleotide that hybridized to the 3′ nucleotide sequence on the target nucleic acid molecule.Also disclosed are kits for performing the methods of the invention.
Owner:SYNGENTA PARTICIPATIONS AG
Assay for localized detection of analytes
InactiveUS20110223585A1Efficient localizationSimple and convenient visualizationMicrobiological testing/measurementAssayAnalyte
The present invention relates to a method for detecting an analyte in a sample, said method comprising: (a) contacting said sample with at least one set of at least first and second proximity probes, wherein said probes each comprise an analyte-binding moiety and can simultaneously bind to the analyte, and wherein (i) said first proximity probe comprises a nucleic acid moiety attached at one end to the analyte-binding moiety, wherein a circular or circularizable oligonucleotide is hybridized to said nucleic acid moiety before, during or after said contacting step; and (ii) said second proximity probe comprises an enzyme moiety, attached to the analyte-binding moiety, capable of directly or indirectly enabling rolling circle amplification (RCA) of the circular or, when it is circularized, of the circularizable oligonucleotide hybridized to the nucleic acid moiety of the first proximity probe, wherein said RCA is primed by said nucleic acid moiety of said first proximity probe; (b) if necessary, circularizing said oligonucleotide, to produce a circularized template for RCA; (c) subjecting said circular or circularized template to RCA, wherein if the enzyme moiety of the second proximity probe in step (a)(ii) is a DNA polymerase, this step does not utilize a free DNA polymerase; and (d) detecting a product of said RCA.
Owner:OLINK AB
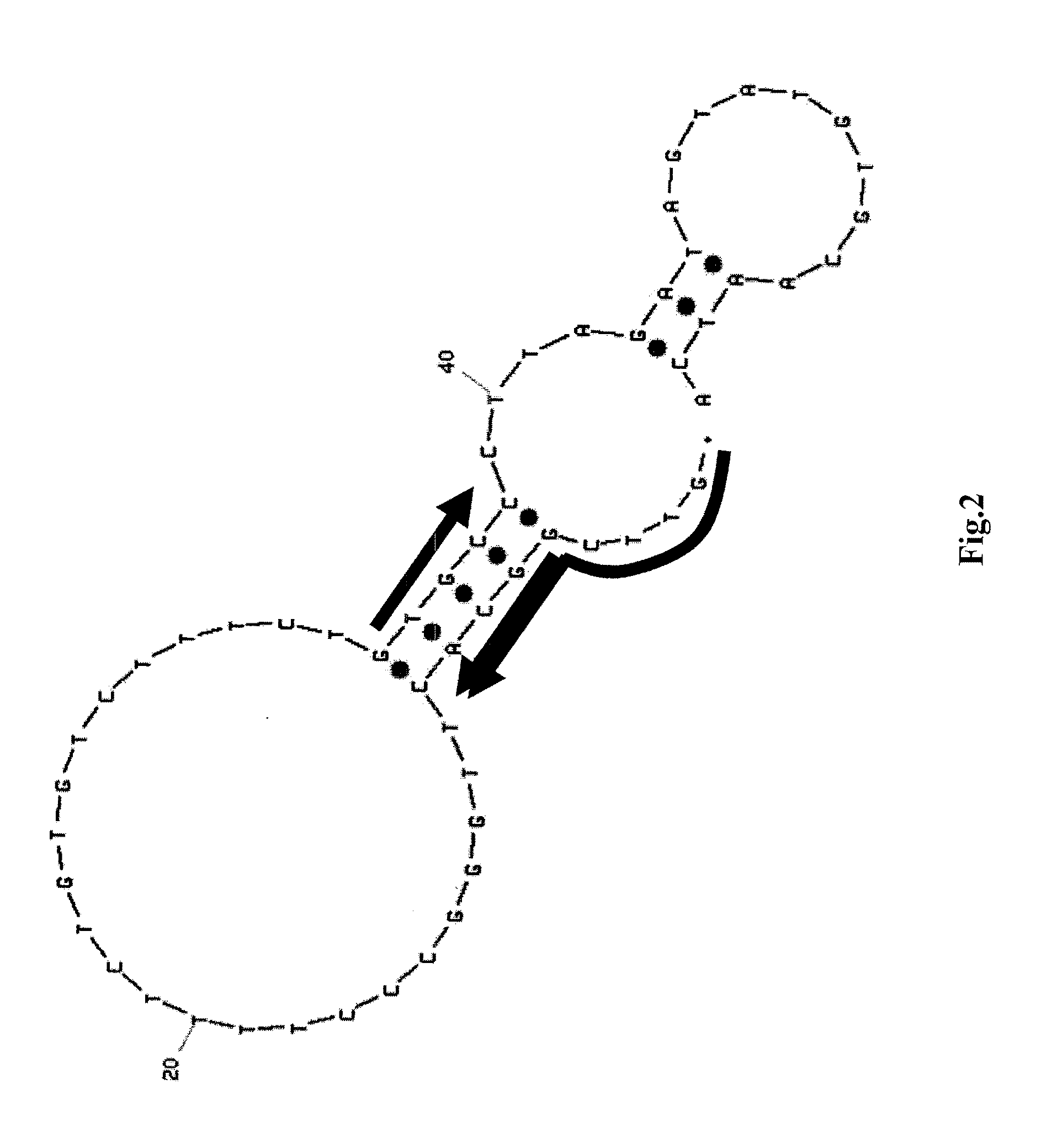
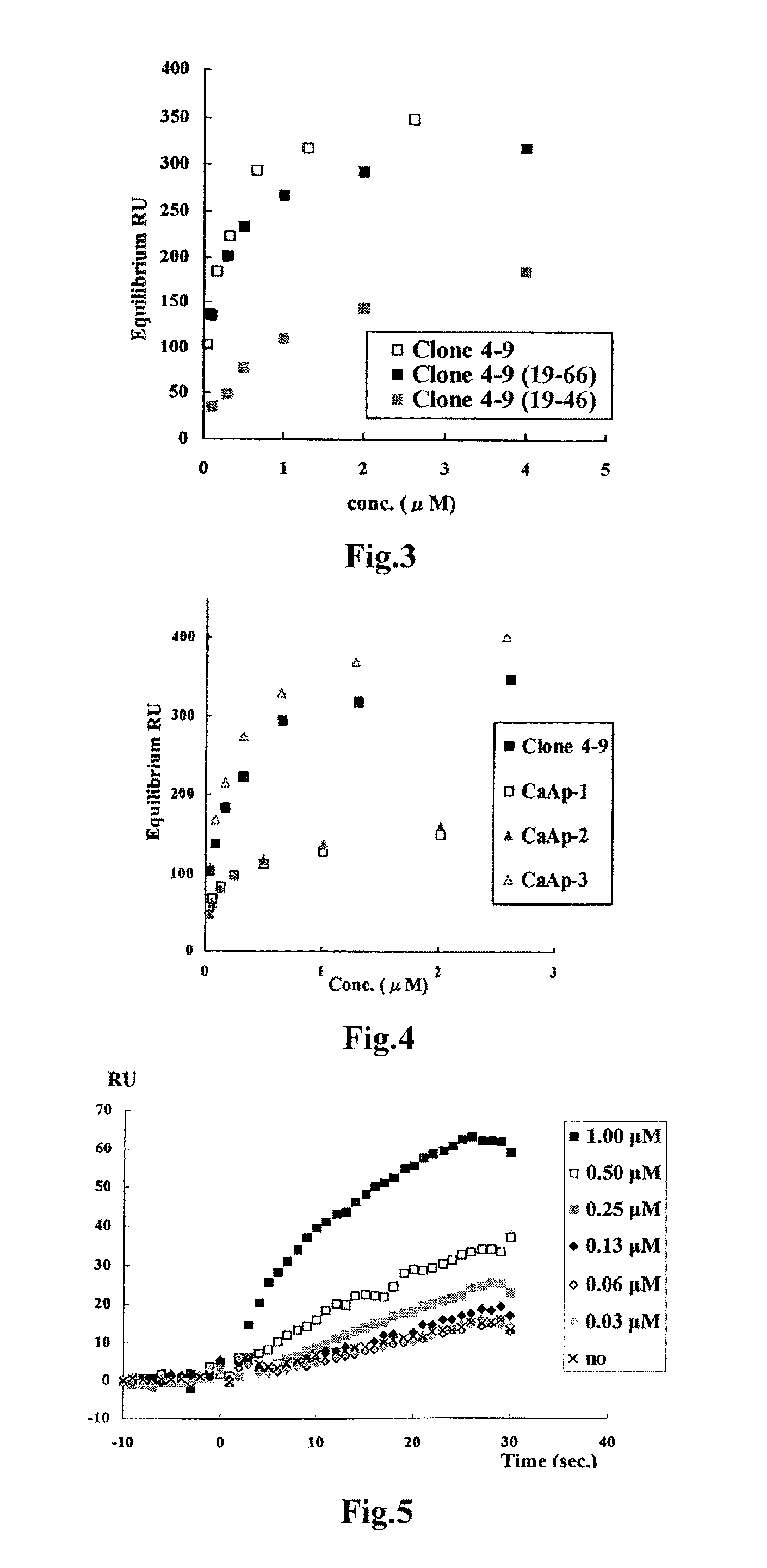
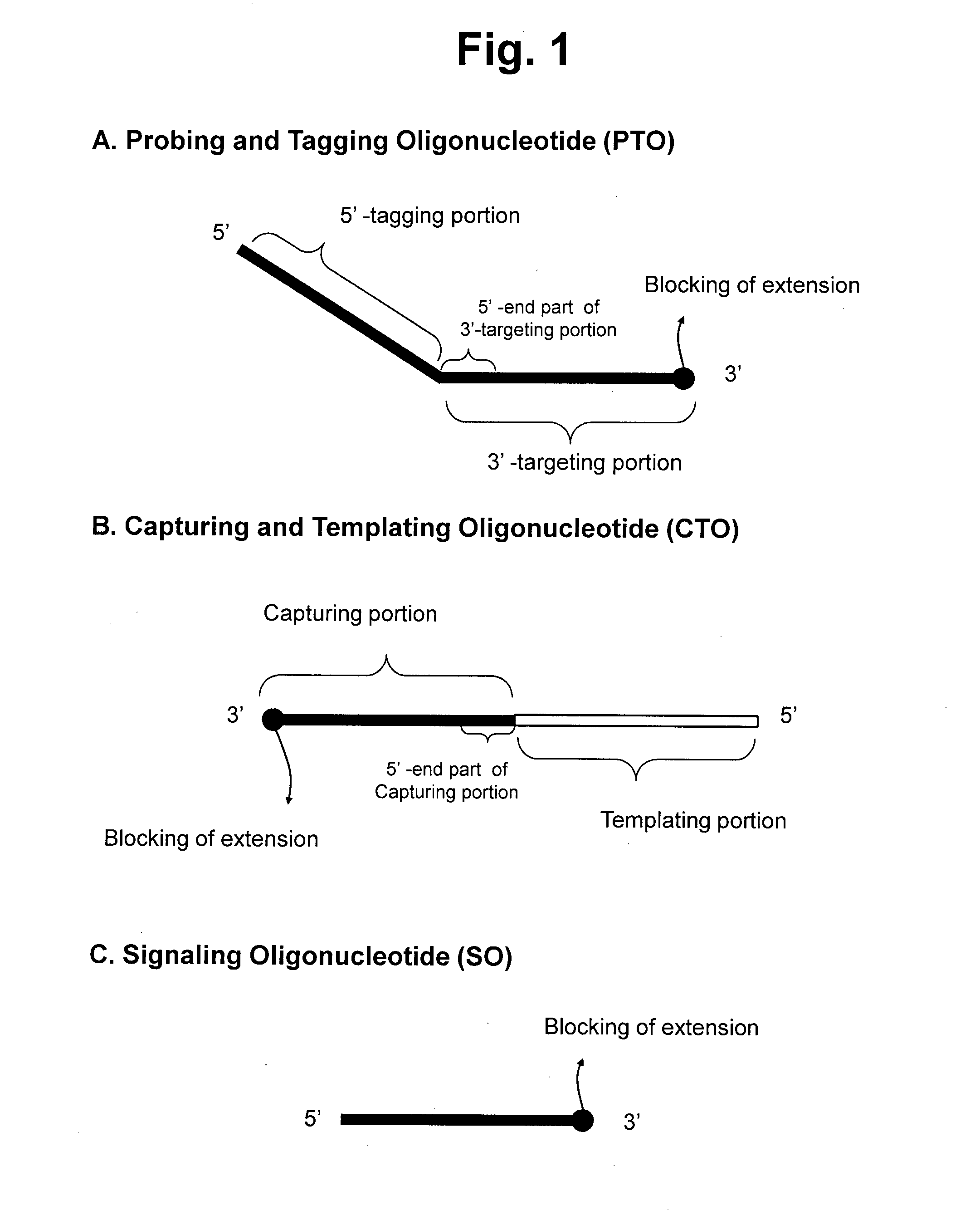
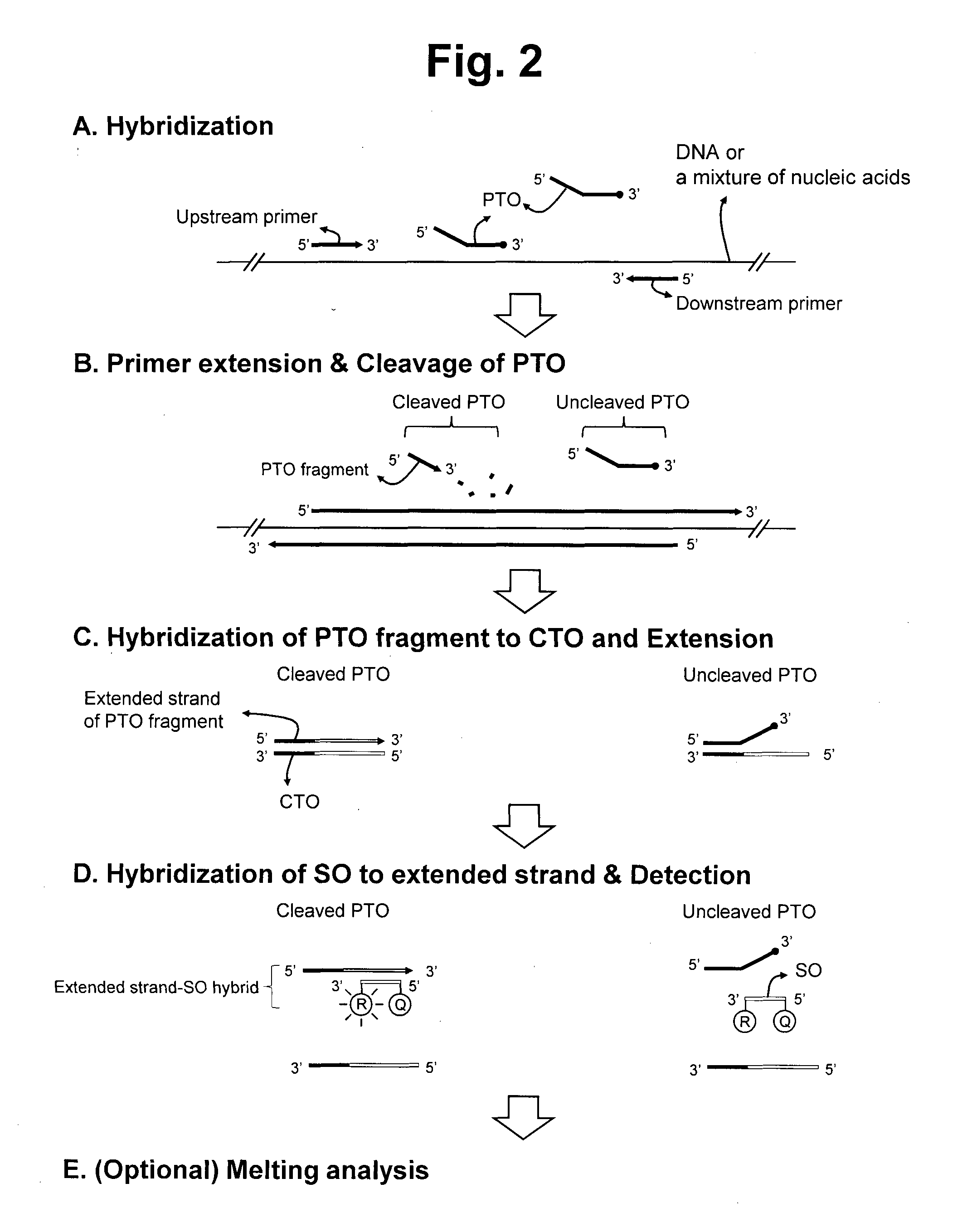
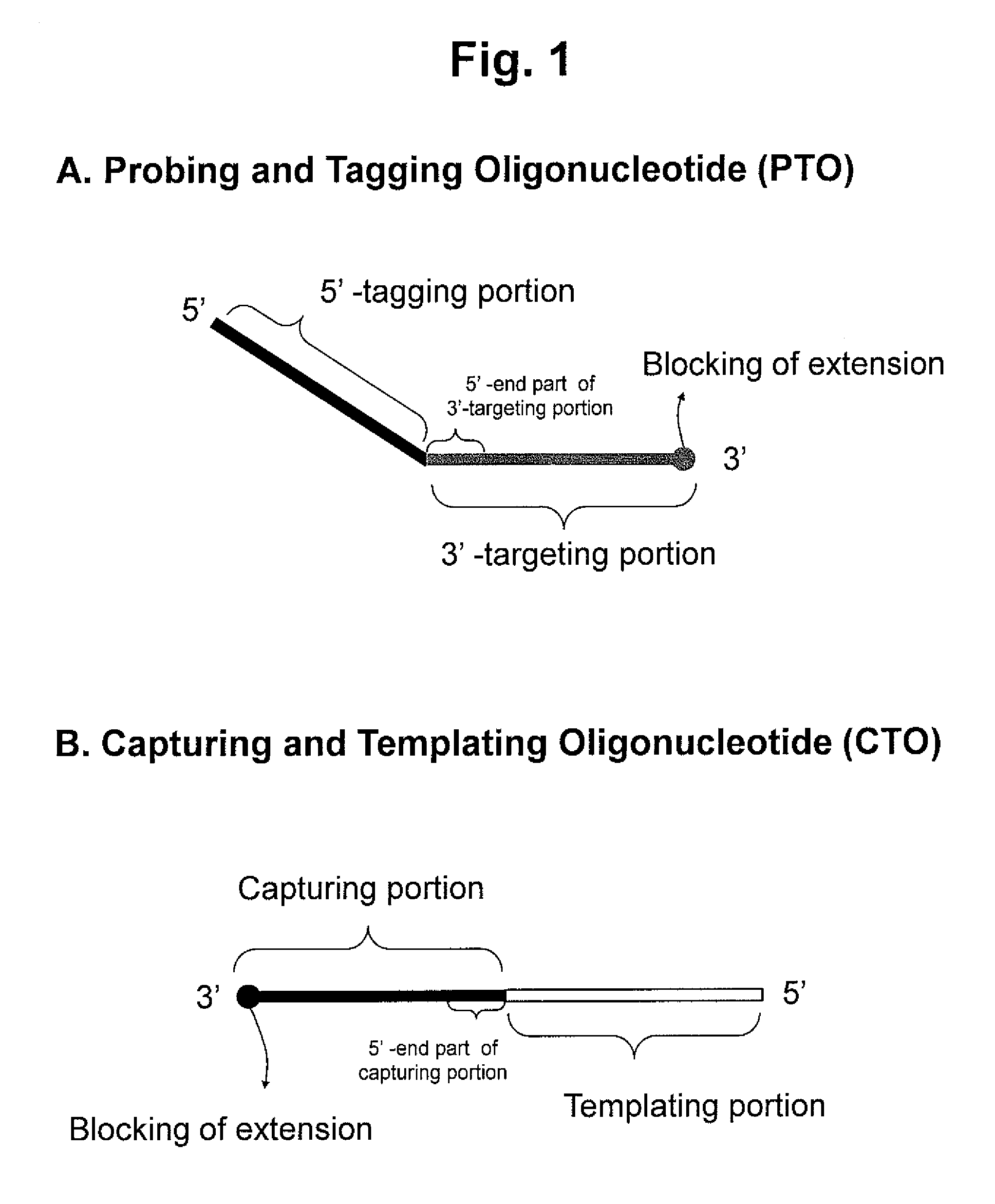
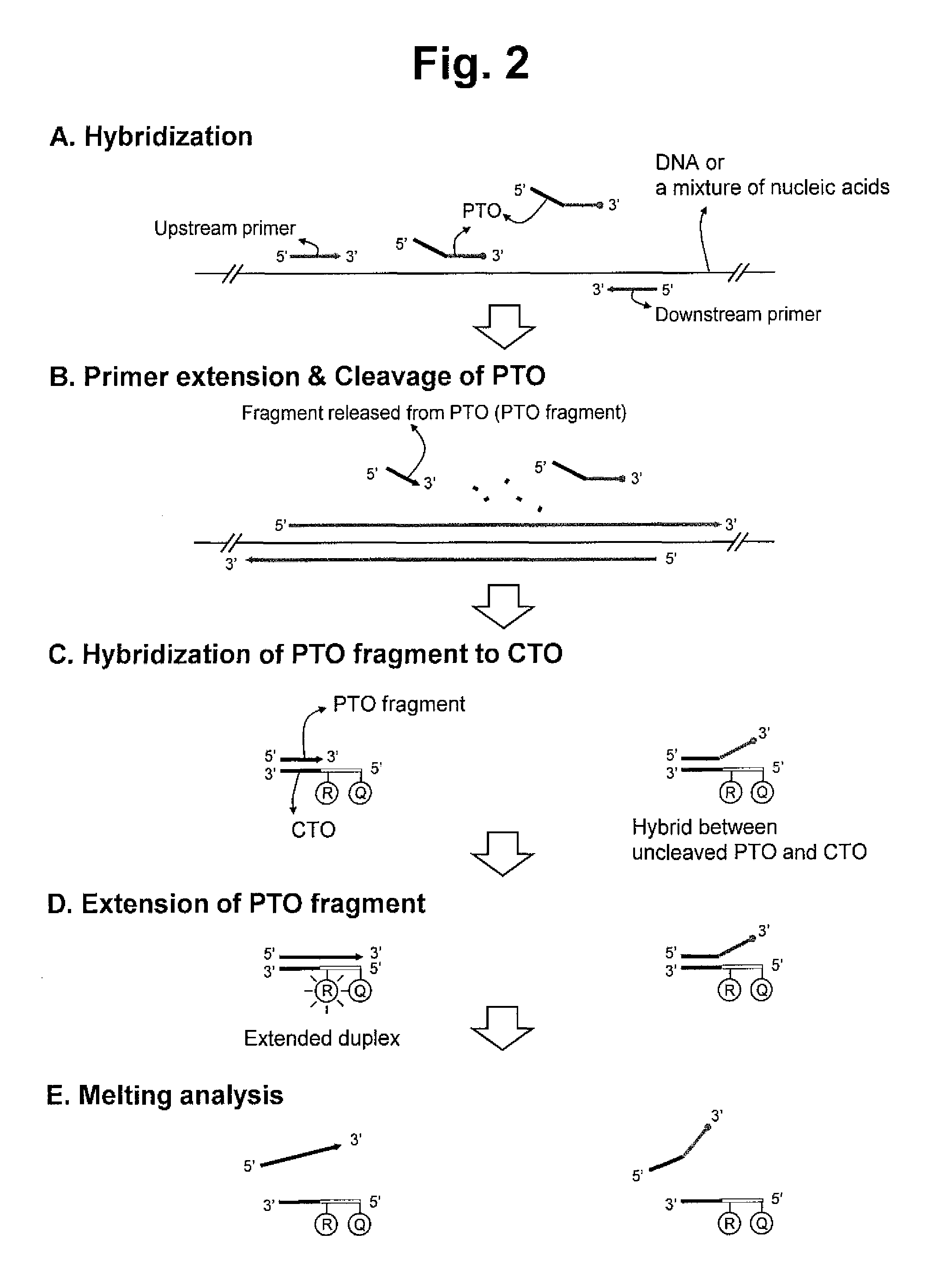
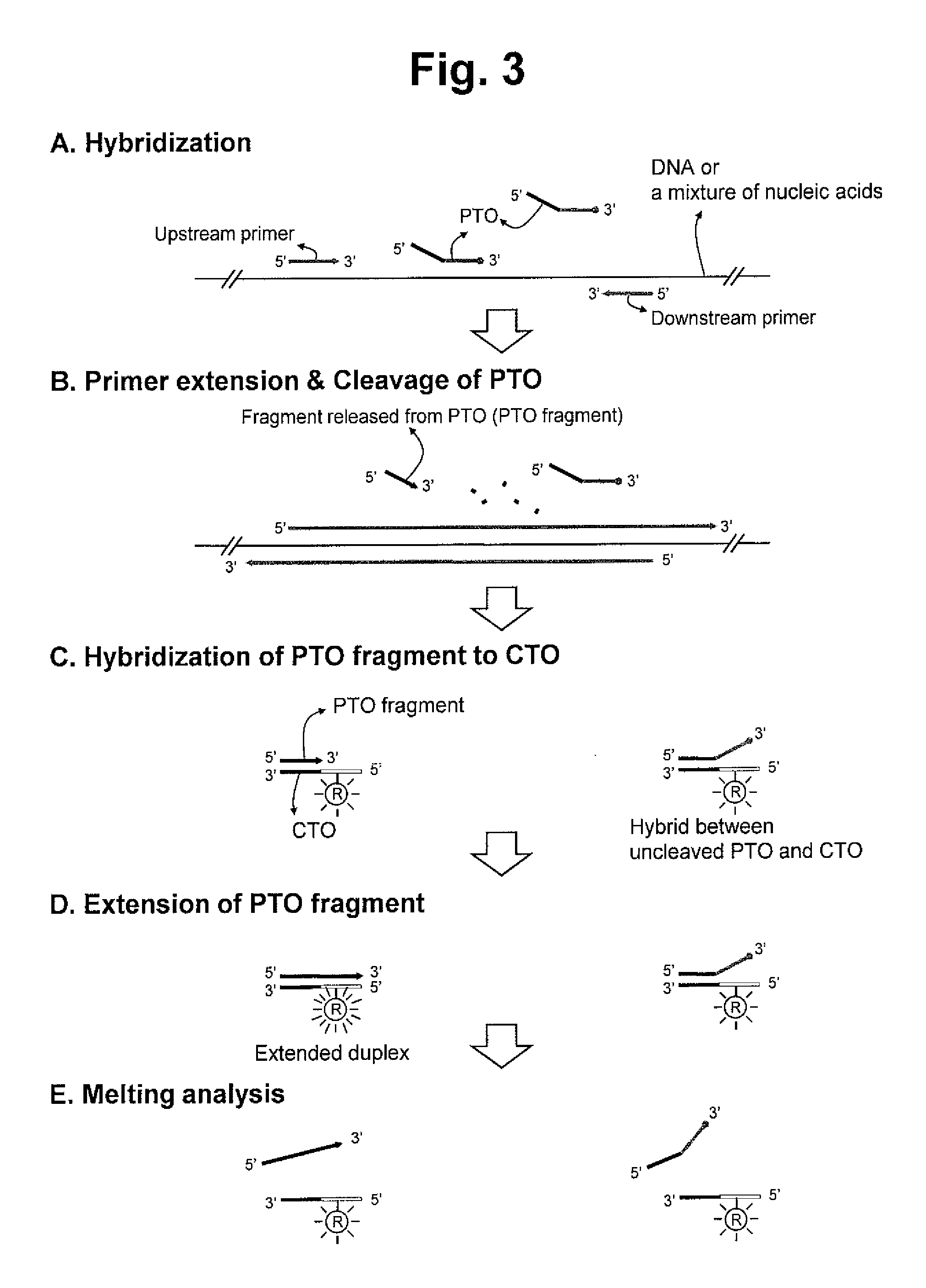
Detection of target nucleic acid sequences by pto cleavage and extension assay
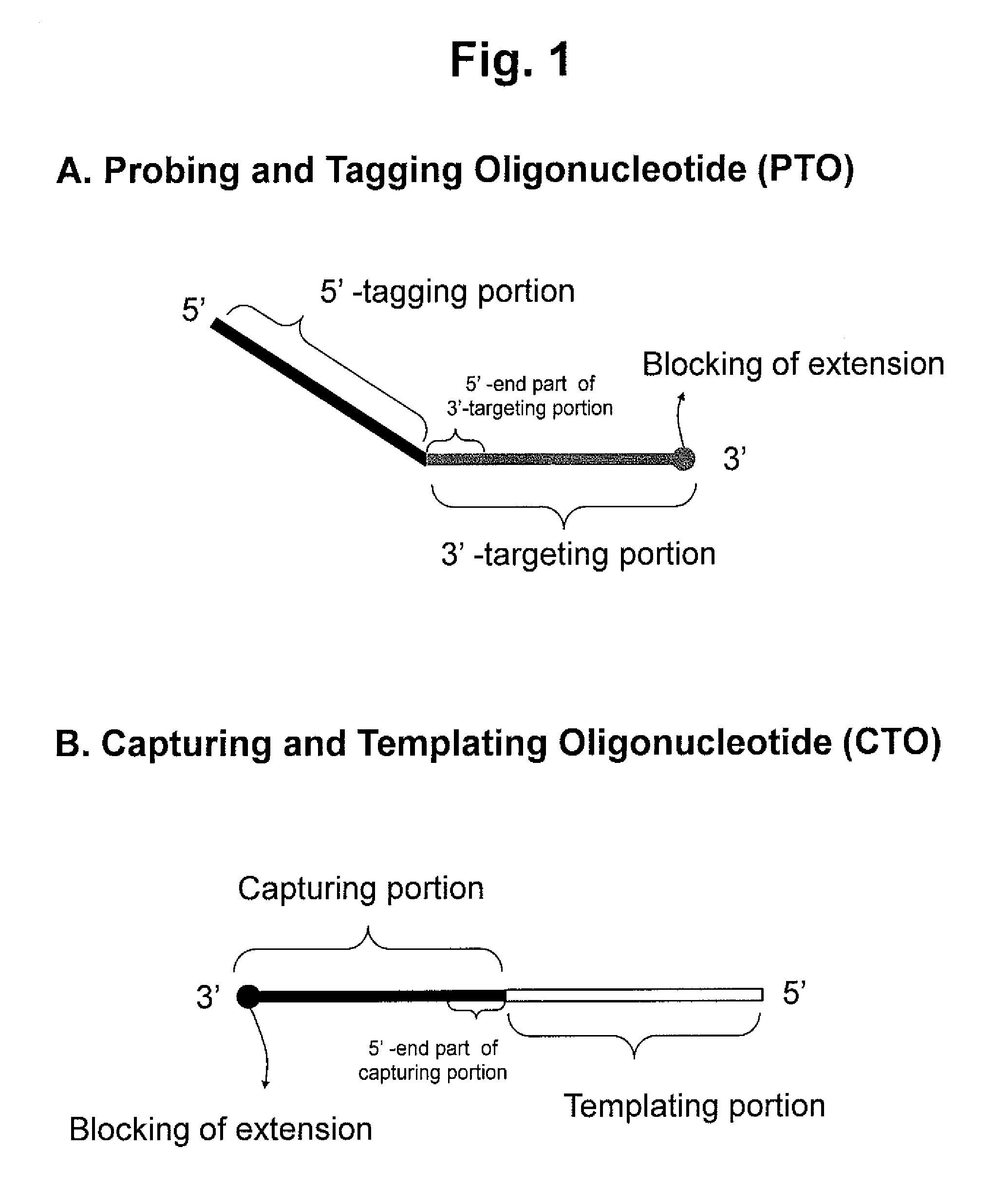
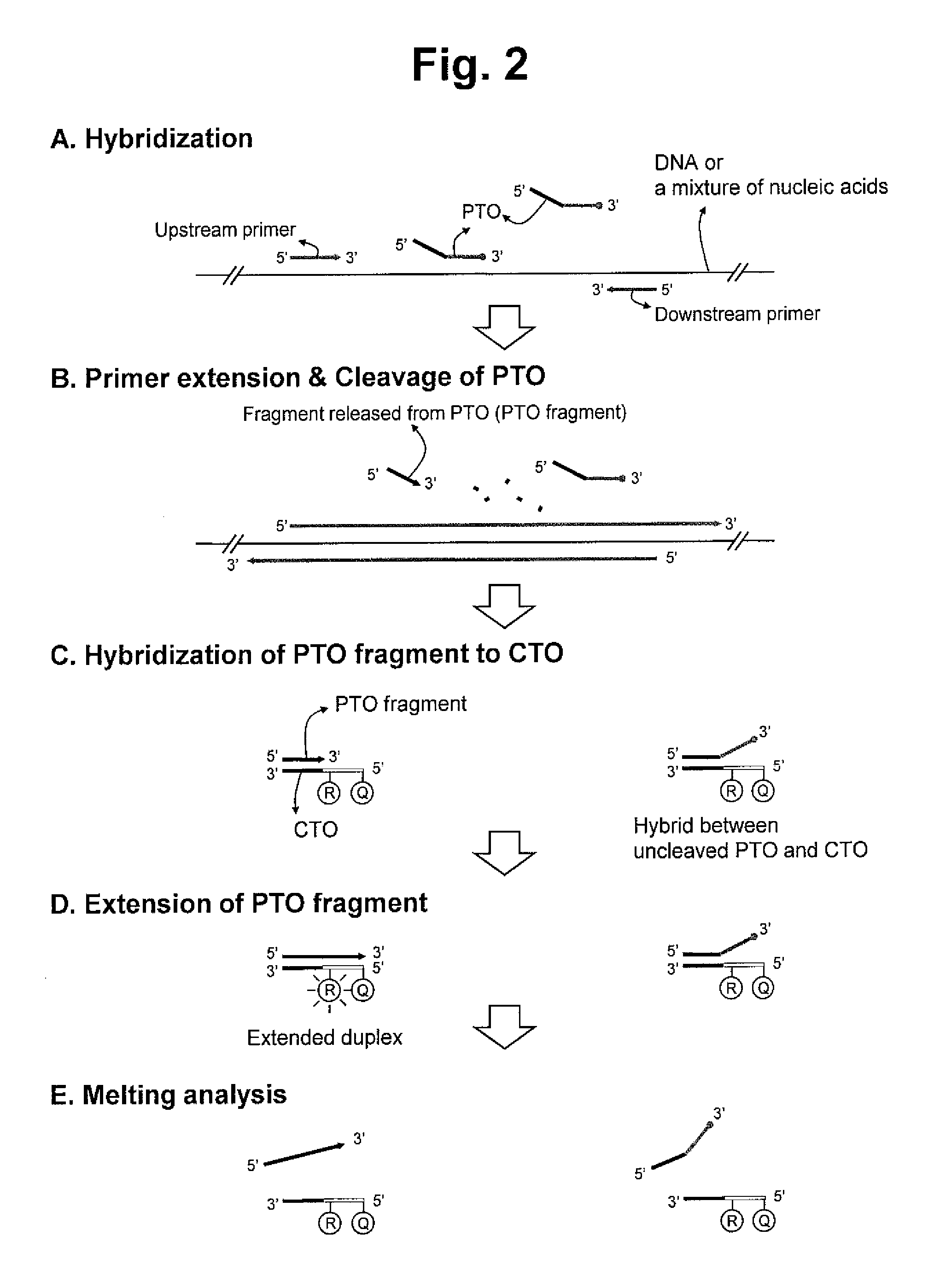
ActiveUS20130109588A1Improve accuracyImprove convenienceMicrobiological testing/measurementLibrary screeningAssayNucleotide
The present invention relates to the detection of a target nucleic acid sequence by a PTOCE (PTO Cleavage and Extension) assay. The present invention detects a target nucleic acid sequence in which the PTO (Probing and Tagging Oligonucleotide) hybridized with the target nucleic acid sequence is cleaved to release a fragment and the fragment is hybridized with the CTO (Capturing and Templating Oligonucleotide) to form an extended duplex, followed by detecting the presence of the extended duplex. The extended duplex provides signals (generation, increase, extinguishment or decrease of signals) from labels indicating the presence of the extended duplex and has adjustable Tm value, which are well adoptable for detection of the presence of the target nucleic acid sequence.
Owner:SEEGENE INC
Methods for quantitative cDNA analysis in single-cell
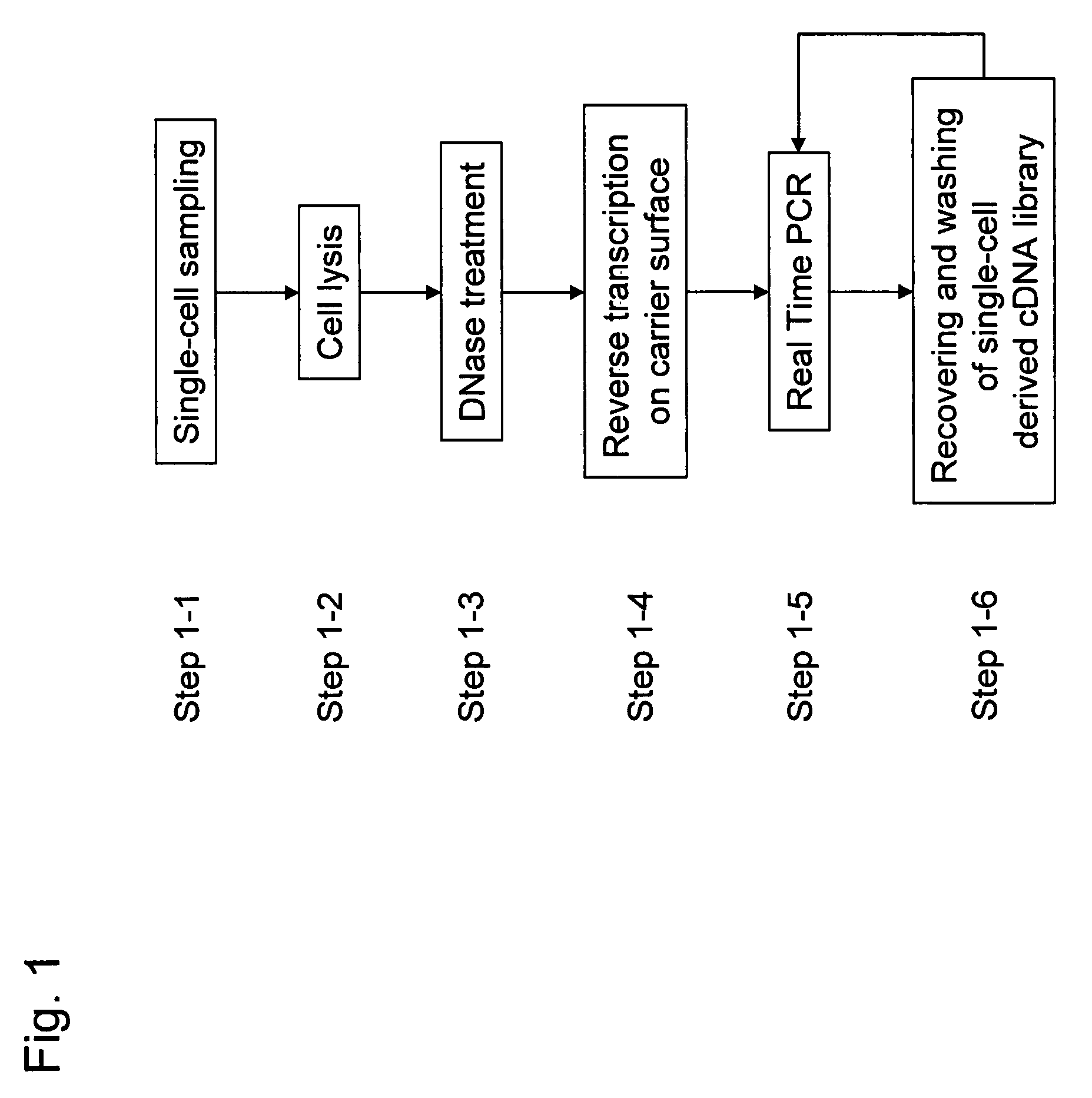
ActiveUS8802367B2Purification of smallSmall recoverySugar derivativesMicrobiological testing/measurementCDNA libraryLysis
It is an object to provide a method of suitably analyzing the amount of gene expression of a single-cell.A method of detecting a nucleic acid comprisinga step of sampling a single-cell from a sample containing at least a single-cell,a cell lysis step of lysing cell membrane of the sampled single-cell and extracting nucleic acids from the cell,a DNase treatment step of degrading DNA of the extracted nucleic acids with DNase,a step of hybridizing mRNA of the total RNA contained in the single-cell with oligo (dT) fixed onto a carrier,a step of performing reverse transcription of the mRNA hybridized with the oligo (dT) to fix cDNA derived from the single-cell onto the carrier, thereby preparing a single-cell derived cDNA library fixed onto a carrier, anda step of amplifying cDNA fixed onto the carrier and simultaneously detecting an amplification amount of the cDNA.
Owner:HITACHI LTD
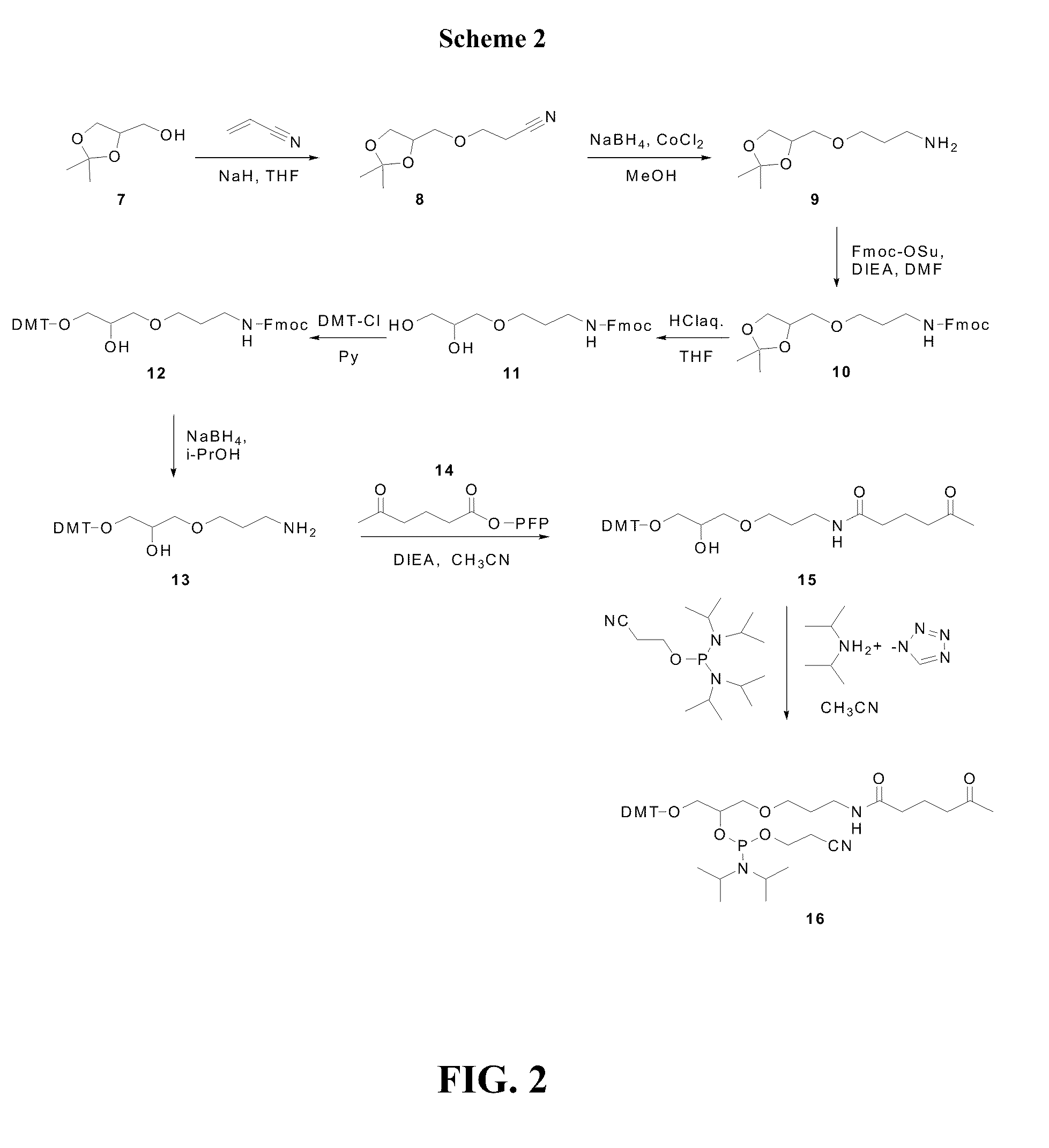
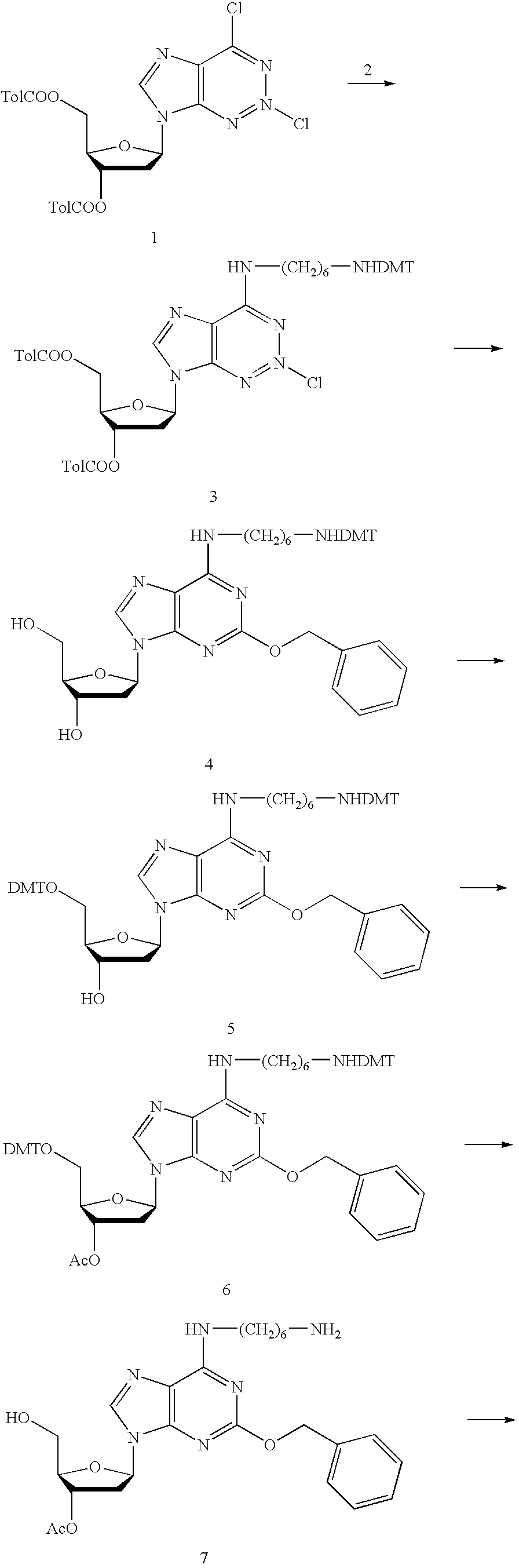
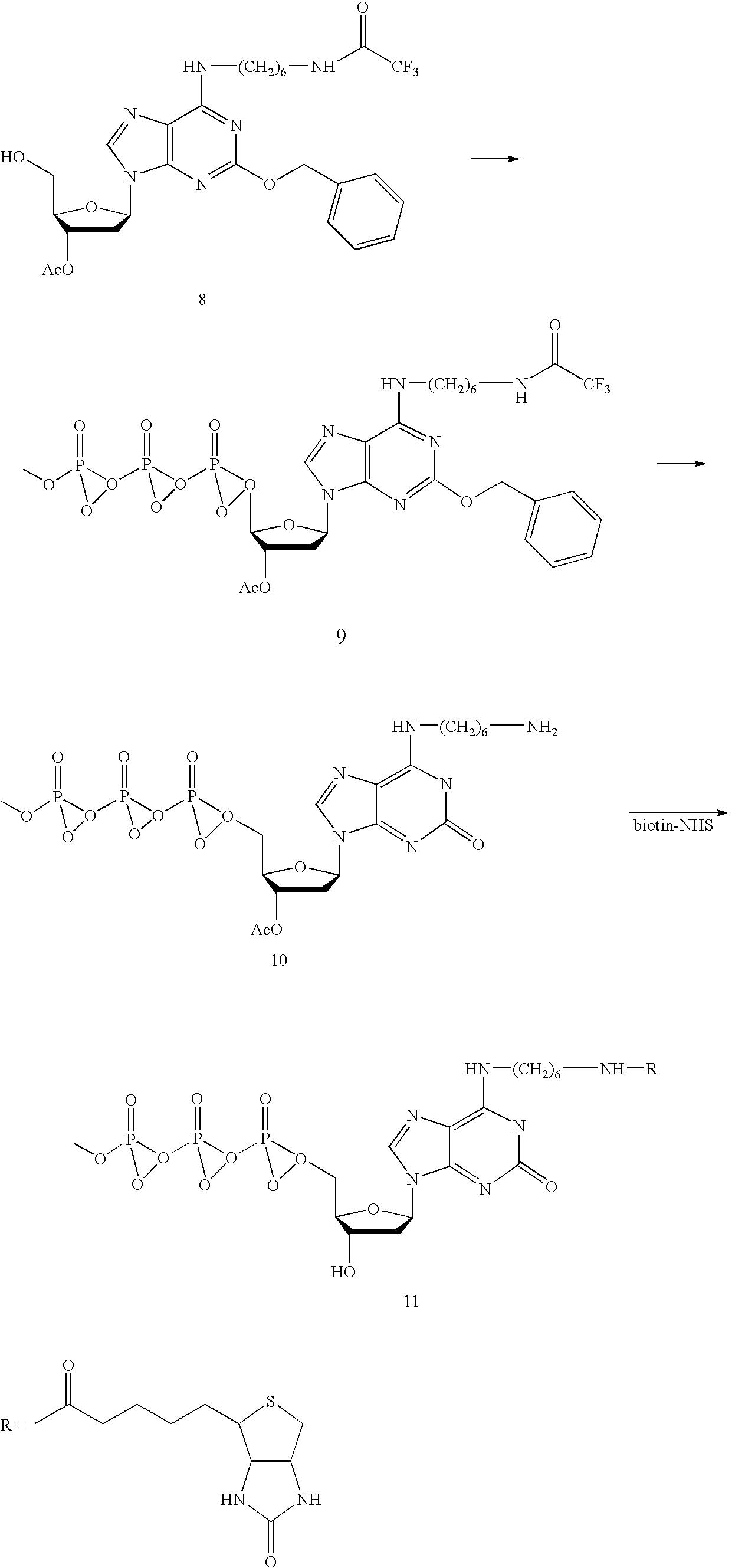
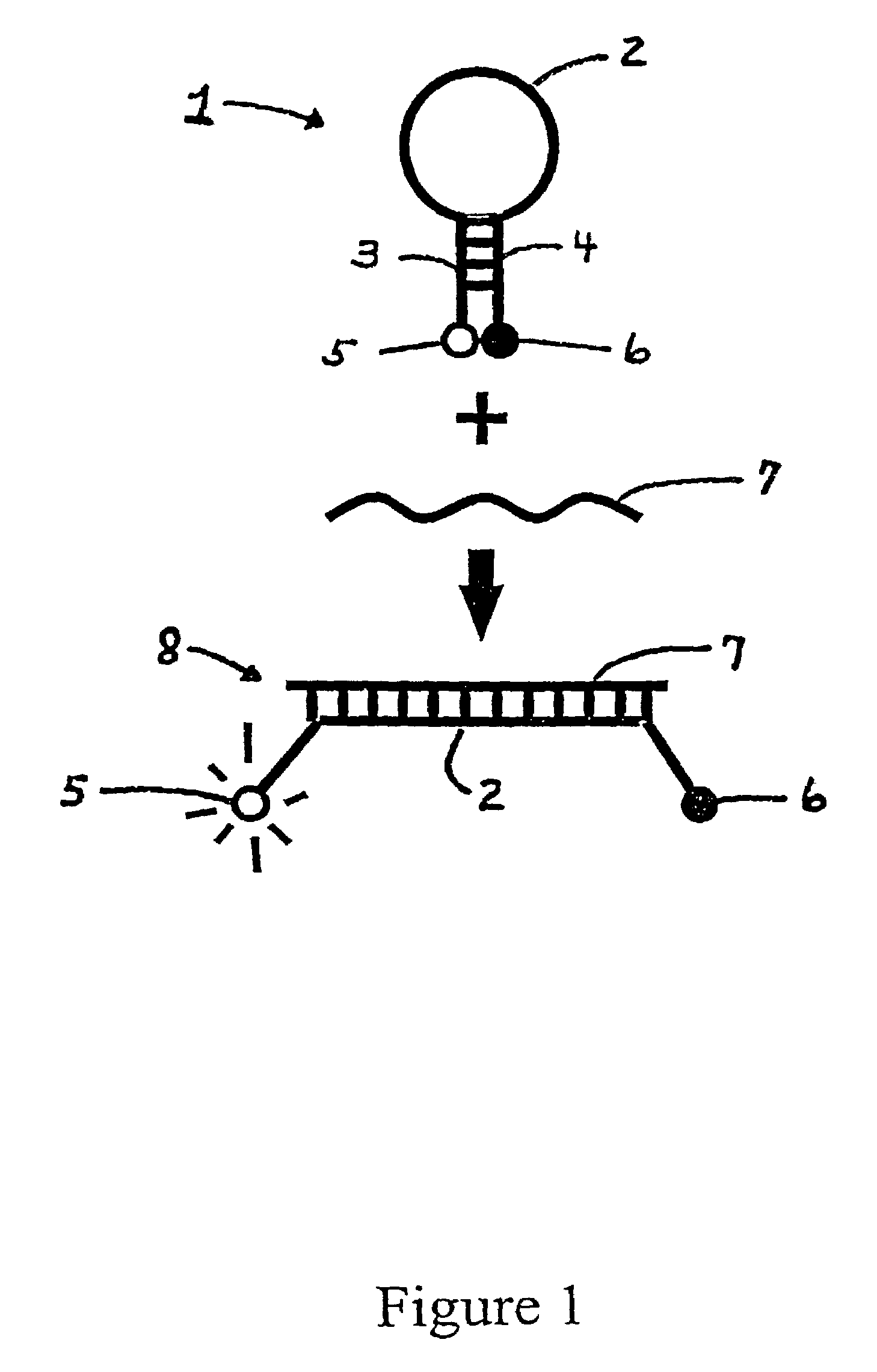
Compounds and methods for labeling oligonucleotides
The invention provides a novel method of labeling oligonucleotides, with reporter moieties, including but not limited to, quenchers, fluorophores, biotin, digoxigenin, peptides and proteins. In addition, this invention provides a method of detecting hybridization of oligonucleotides. This invention also provides novel azo quenchers having the general formula shown below.The invention further provides compositions comprising labeled oligonucleotides and solid supports. The invention also provides kits comprising at least one composition of the present invention.
Owner:INTEGRATED DNA TECHNOLOGIES
Method to detect prostate cancer in a sample
InactiveUS20050282170A1Easy to separateConsistent resultMicrobiological testing/measurementNucleotideNucleic acid sequencing
The present invention relates to prostate cancer. More specifically, the present invention relates to a method to detect prostate cancer in a patient sample by detecting the RNA encoded by the gene PCA3. More particularly the present invention relates to a method for determining a predisposition, or presence of prostate cancer in a patient comprising: (a) contacting a biological sample of a patient with at least one oligonucleotide that hybridizes to a PCA3 polynucleotide; (b) detecting in the biological sample an amount of PCA3 and second prostate specific polynucleotides; and (c) comparing the amount of PCA3 polynucleotide that hybridizes to the oligonucleotide to a predetermined cut off value, and therefrom determining the presence or absence of prostate cancer in the biological sample. The present invention further relates to diagnostic kits for the detection of prostate cancer or the risk of developing same in a patient comprising: (a) at least one container means having disposed therein at least one oligonucleotide probe or primer that hybridizes to one a PCA3 nucleic acid or complement thereof; (b) at least one oligonucleotide probe or primer that hybridizes with a second prostate specific nucleic acid or complement thereof; and (c) reagents enabling a detection of PCA3 and of the second prostate specific nucleic acid when PCA3 or second prostate-specific nucleic acid sequence is present.
Owner:GEN PROBE INC
Detection of nucleic acids
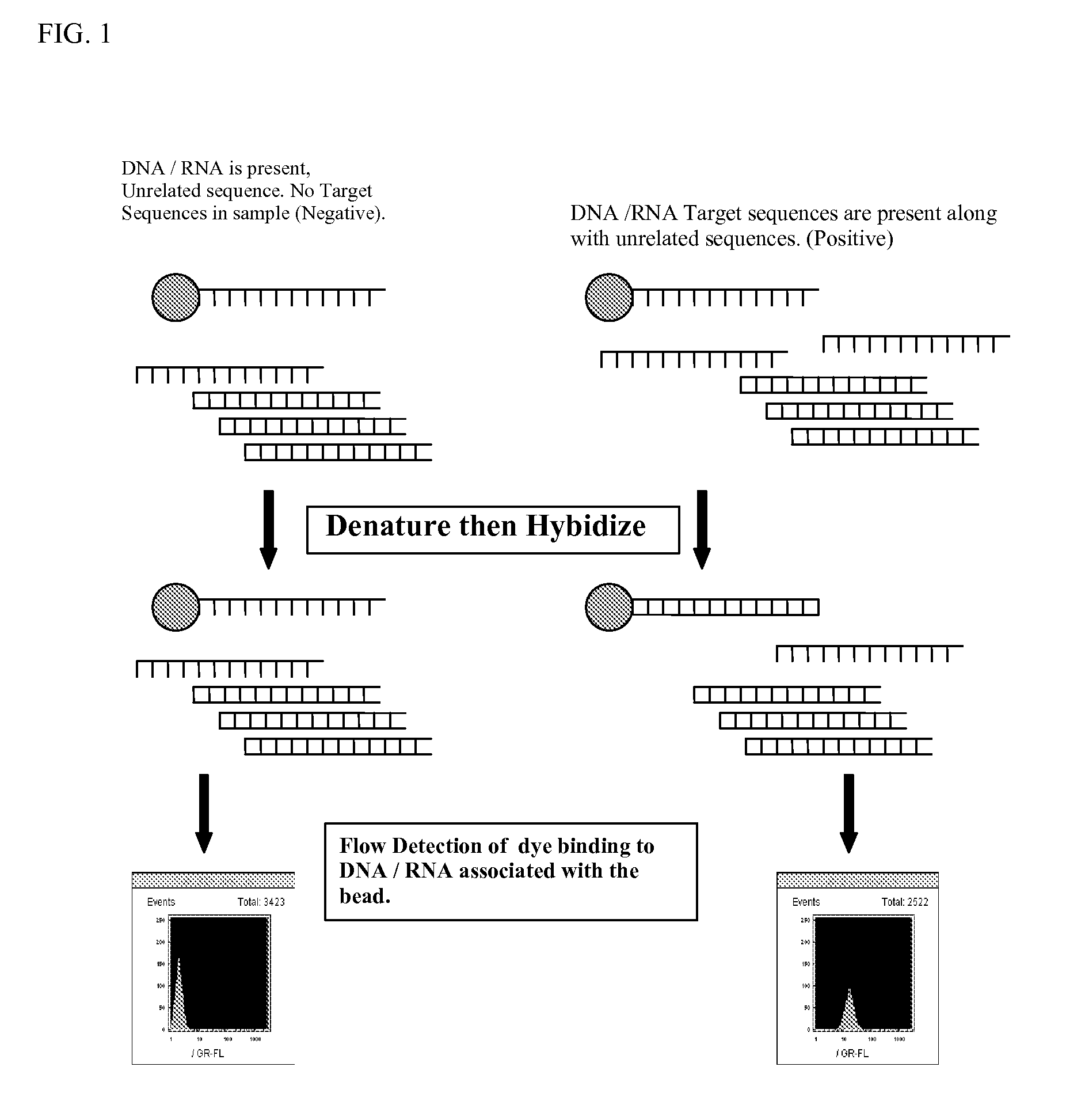
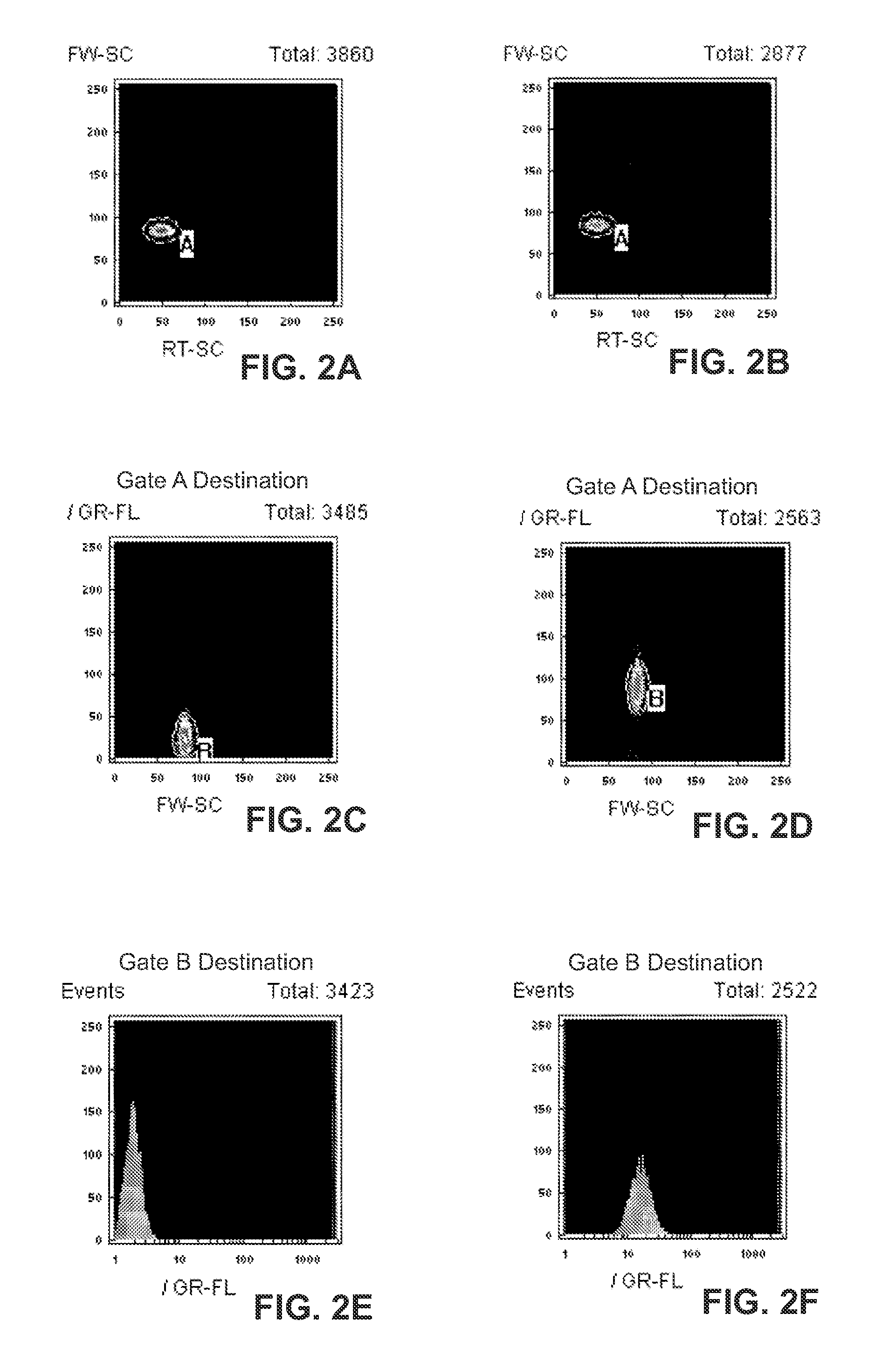
InactiveCN1312287CBioreactor/fermenter combinationsAnalysis using chemical indicatorsParticulatesFluorescence
Homogeneous and multiplex assays for target nucleic acids hybridized to oligonucleotide probes immobilized on particulate and non-particulate substrates are described. Target nucleic acids are determined using detection methods, such as flow cytometric methods, capable of particle discrimination based on the light scattering or fluorescence properties of the particle, or by spatial resolution of oligonucleotides immobilized at specific loci on a substrate. Target-correlated fluorescence signal, originating from a target nucleic acid hybridized to the substrate-immobilized oligonucleotide is determined as a measure of the presence or amount of the target nucleic acid.
Owner:ORTHO-CLINICAL DIAGNOSTICS
Compounds and methods for labeling oligonucleotides
Owner:INTEGRATED DNA TECHNOLOGIES
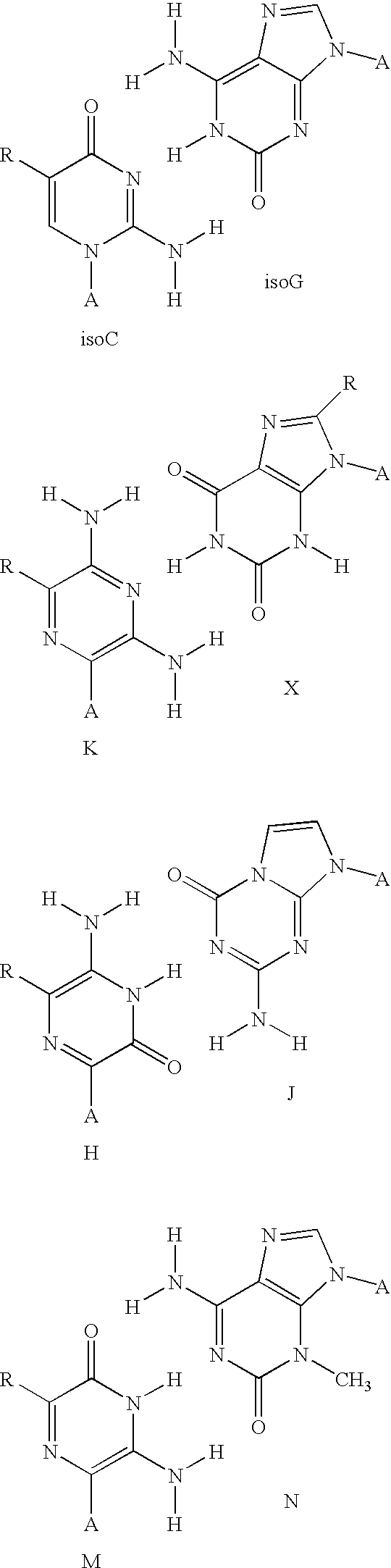
Solid support assay systems and methods utilizing non-standard bases
Solid support assays using non-standard bases are described. A capture oligonucleotide comprising a molecular recognition sequence is attached to a solid support and hybridized with a target oligonucleotide. In some instances, the molecular recognition sequence includes one or more non-standard bases and hybridizes to a complementary tagging sequence of the target oligonucleotide. In other instances, incorporation of a non-standard base (e.g., via PCR or ligation) is used in the assay.
Owner:LUMINEX
Assays for short sequence variants
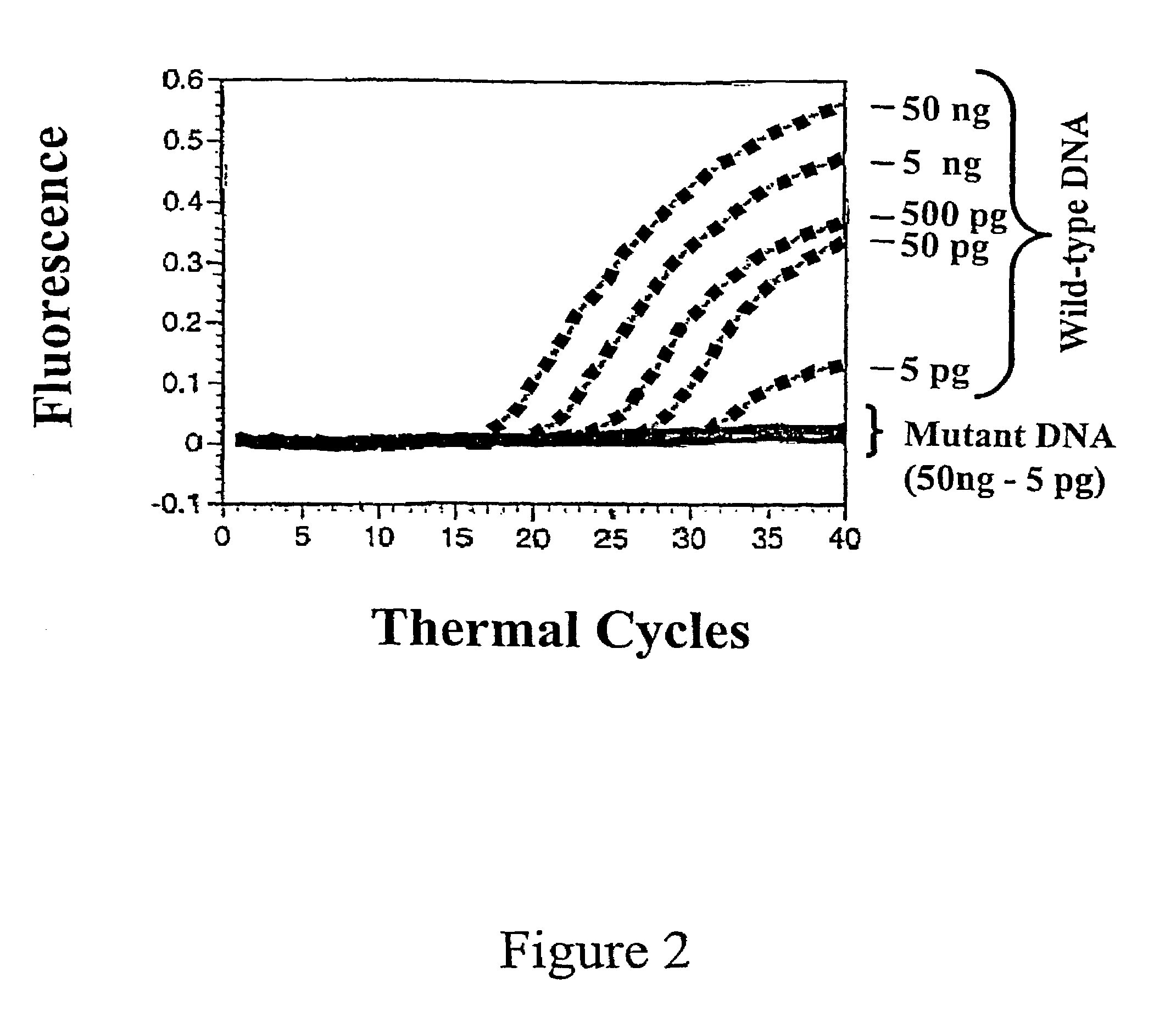
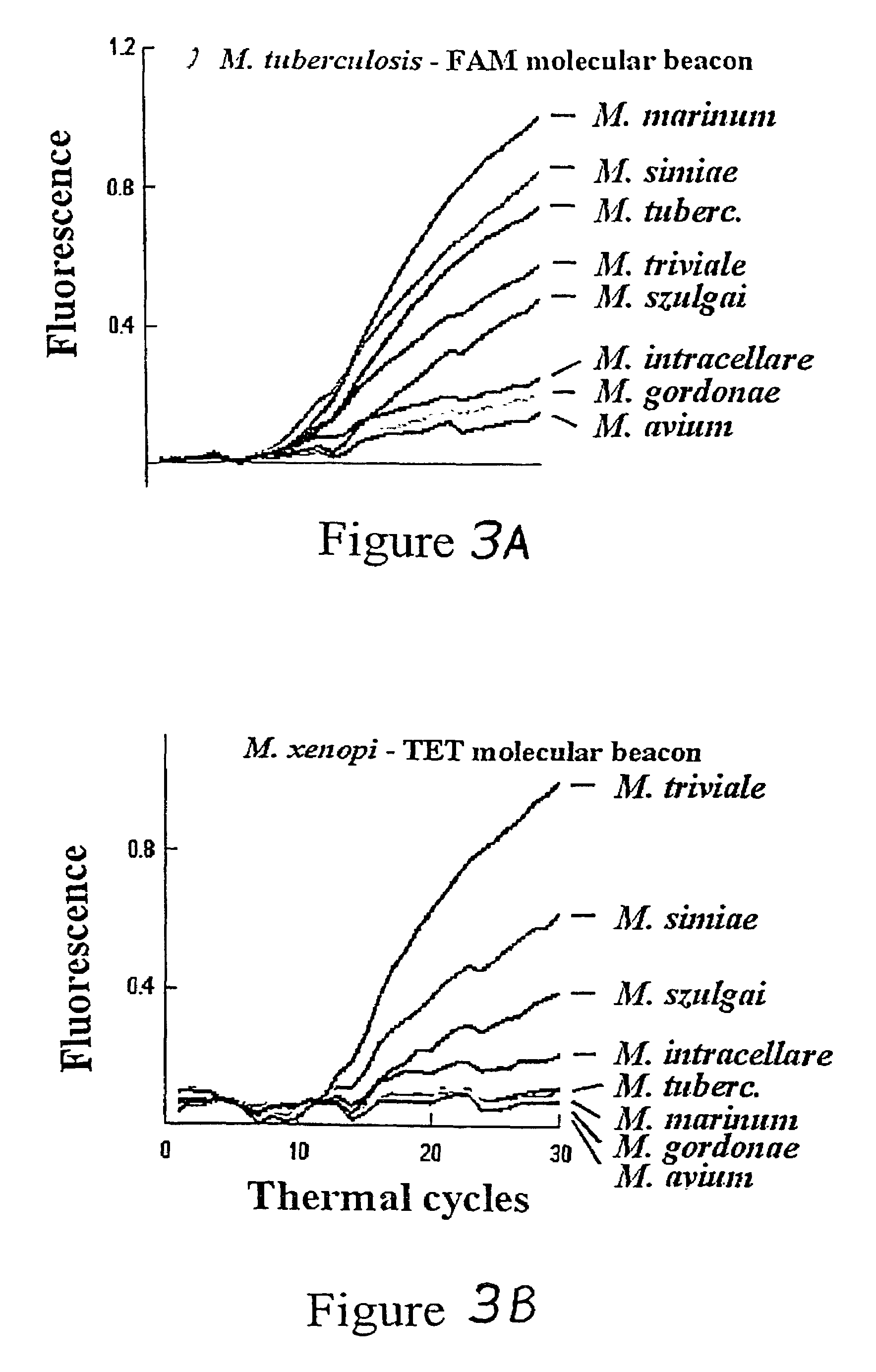
InactiveUS7662550B1Reduce in quantityInexpensive and rapid diagnosticSugar derivativesMicrobiological testing/measurementBioinformaticsGenetic variants
The invention provides assays that can detect multiple genetic variants of a gene (e.g., a mycobacterial gene) in a sample using a pool (e.g., 2, 3, 4, or more) of oligonucleotide hybridization probes.
Owner:PHRI PROPERTIES
Method of assaying target substance in sample, aptamer molecule and method of constructing the same
InactiveUS20100021899A1Easy and inexpensiveCarry-out inexpensivelySugar derivativesMicrobiological testing/measurementNucleotideAmount of substance
A method for measuring a test substance whereby the test substance in a sample can be specifically measured without using an antibody against the test substance; an aptamer molecule used therefor; and a method for creating the aptamer are disclosed. An aptamer capable of hybridizing with an oligonucleotide when it is bound to a test substance, but is incapable of hybridizing with the oligonucleotide when it is not bound to the test substance, is utilized. The aptamer is brought into contact with a sample, and the aptamer bound to the test substance is brought into contact with an immobilized oligonucleotide which hybridizes with the aptamer, to bind the aptamer to a solid phase, followed by measurement of the aptamer immobilized on the solid phase.
Owner:NAT UNIV CORP TOKYO UNIV OF AGRI & TECH
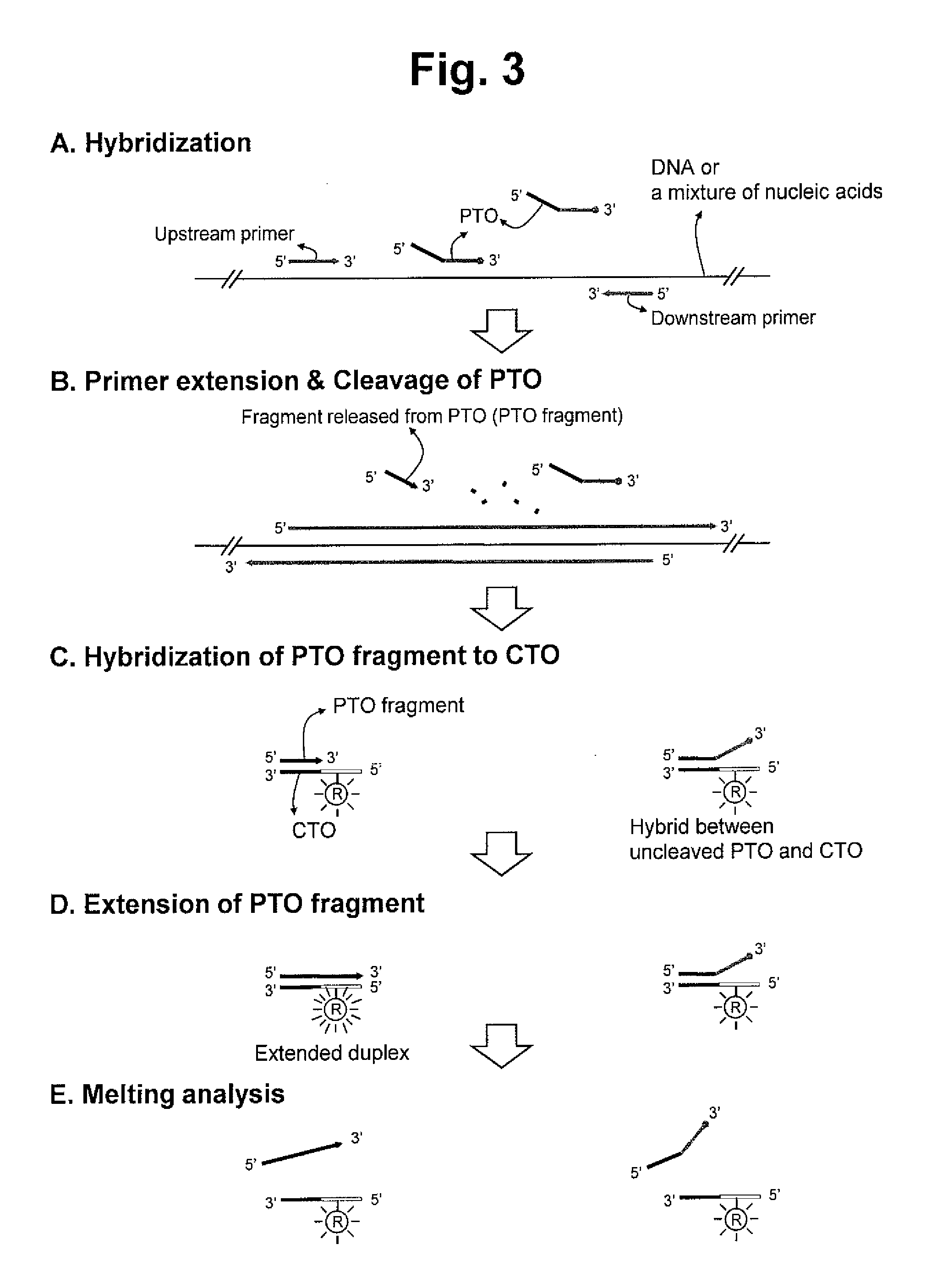
Detection of target nucleic acid sequence by pto cleavage and extension-dependent signaling oligonucleotide hybridization assay
ActiveUS20150072887A1Improve accuracyImprove convenienceNucleotide librariesMicrobiological testing/measurementTarget signalNucleic acid sequencing
The present invention relates to the detection of a target nucleic acid sequence by a PCE-SH (PTO Cleavage and Extension-Dependent Signaling Oligonucleotide Hybridization) assay. The present invention does not use probes to be hybridized with target nucleic acid sequences for providing target signals. Interestingly, the present invention uses probes (signaling oligonucleotides) to be hybridized with the extended strand formed in a target-dependent manner in which the extended strand is synthesized using the CTO artificially selected as templates.
Owner:SEEGENE INC
Allele variant detection method, kit and composition
InactiveCN103215361AImprove efficiencyGrowth inhibitionMicrobiological testing/measurementDNA/RNA fragmentationSpecific detectionFluorescence
The invention provides a method for carrying out allele variant specific detection by utilizing an inhibitor, a kit and a composition. The method provided by the invention comprises the following steps of: adding a primer pair and an inhibitor the terminal 3' of which can not be extended by polymerase in a hybridization process into a nucleic acid sample, wherein the primer pair comprises a specific primer and a reverse primer; hybridizing the specific primer and the reverse primer respectively with target sequence mutant type oligonucleotide in the nucleic acid sample to produce an amplicon; hybridizing the inhibitor with a target sequence wild type oligonucleotide in the nucleic acid sample; and carrying out real-time specific detection on a mutant type allele by utilizing a fluorescent dye or a detection pointer. The added inhibitor can not extend in an amplification process, and amplification of a wild type allele variant is inhibited, so that the amplification efficiency of a mutant type allele variant is improved, and the sensitivity and specificity of detection are improved.
Owner:SHENZHEN UNI MEDICA TECH
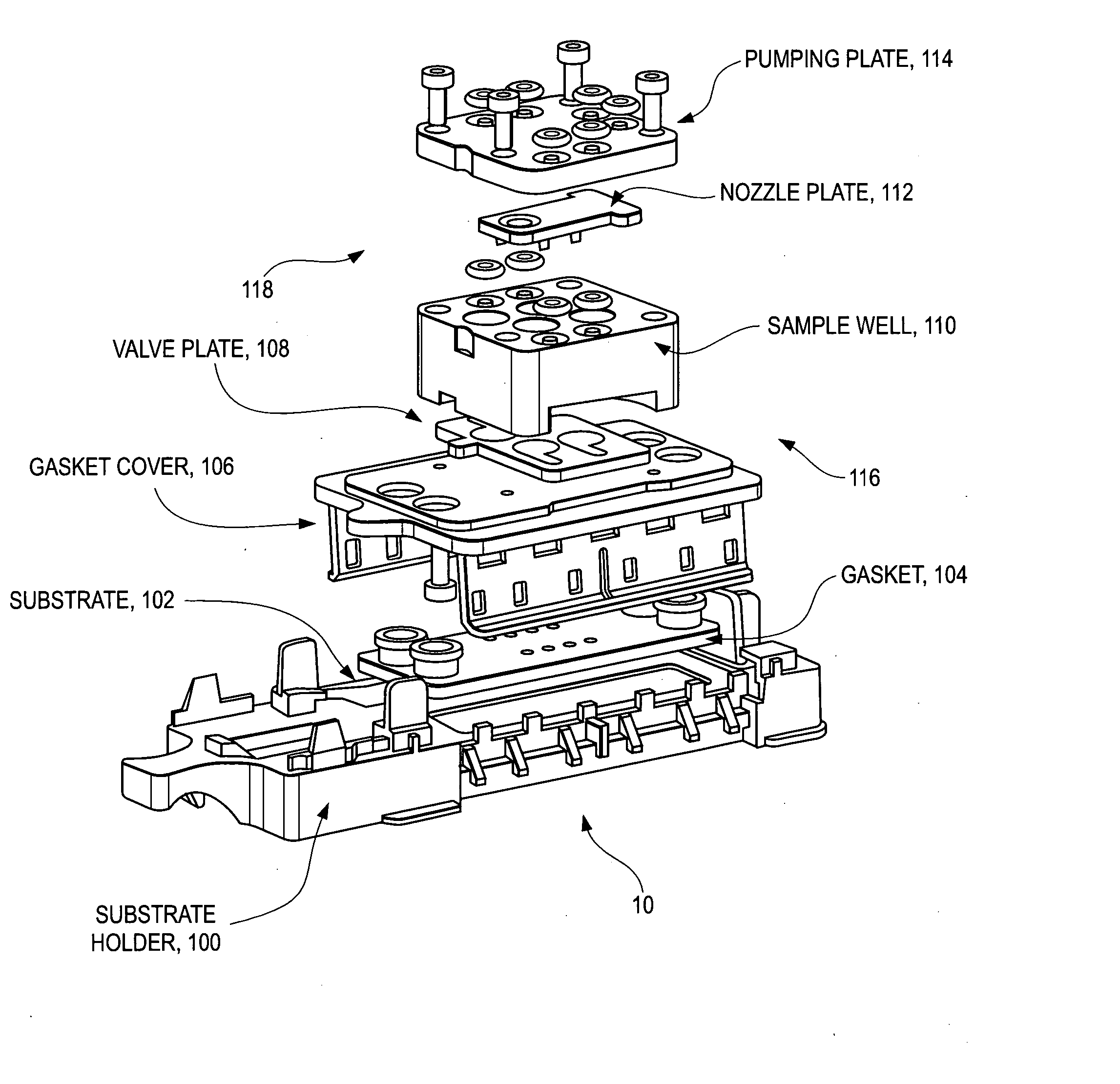
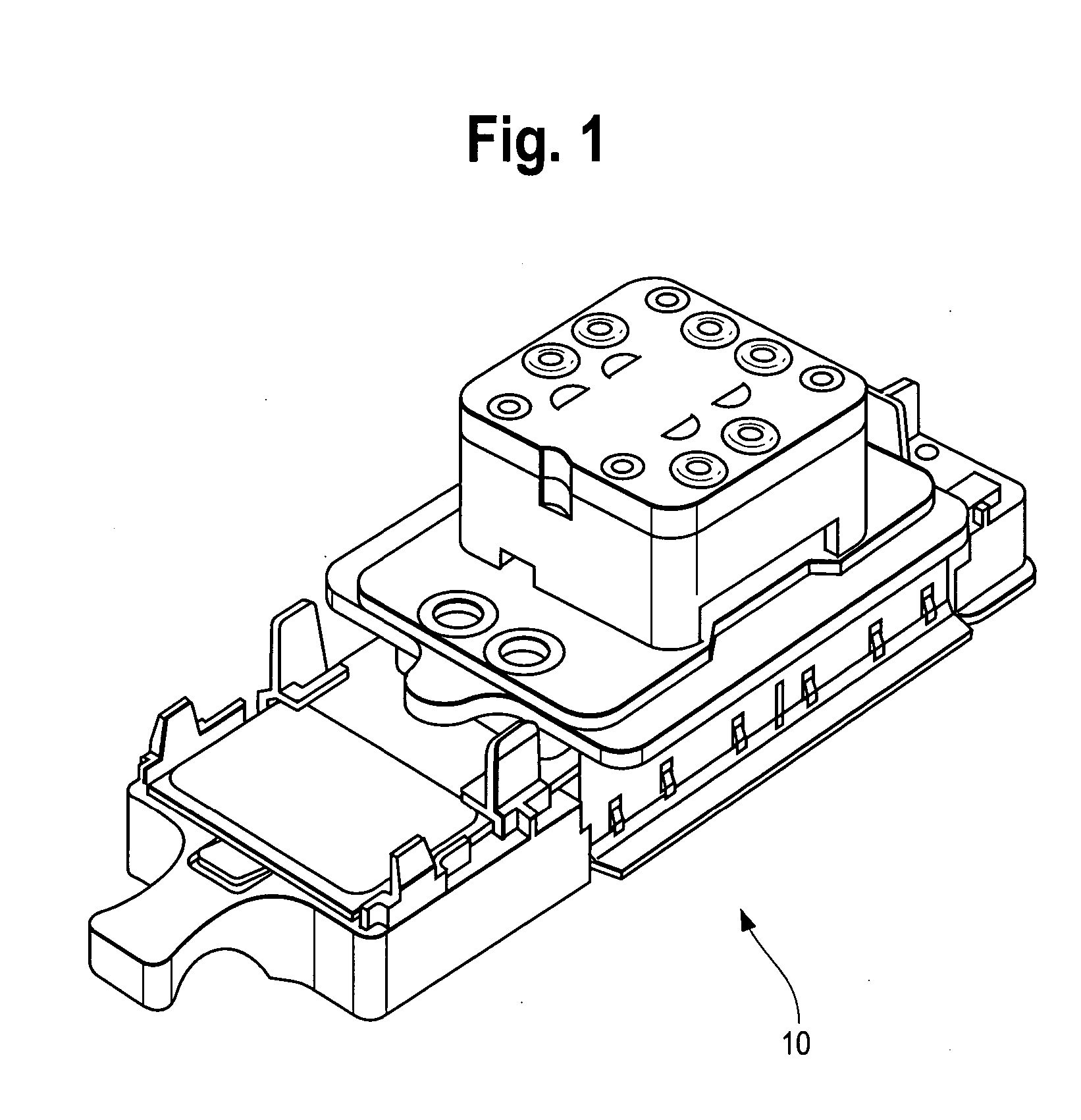
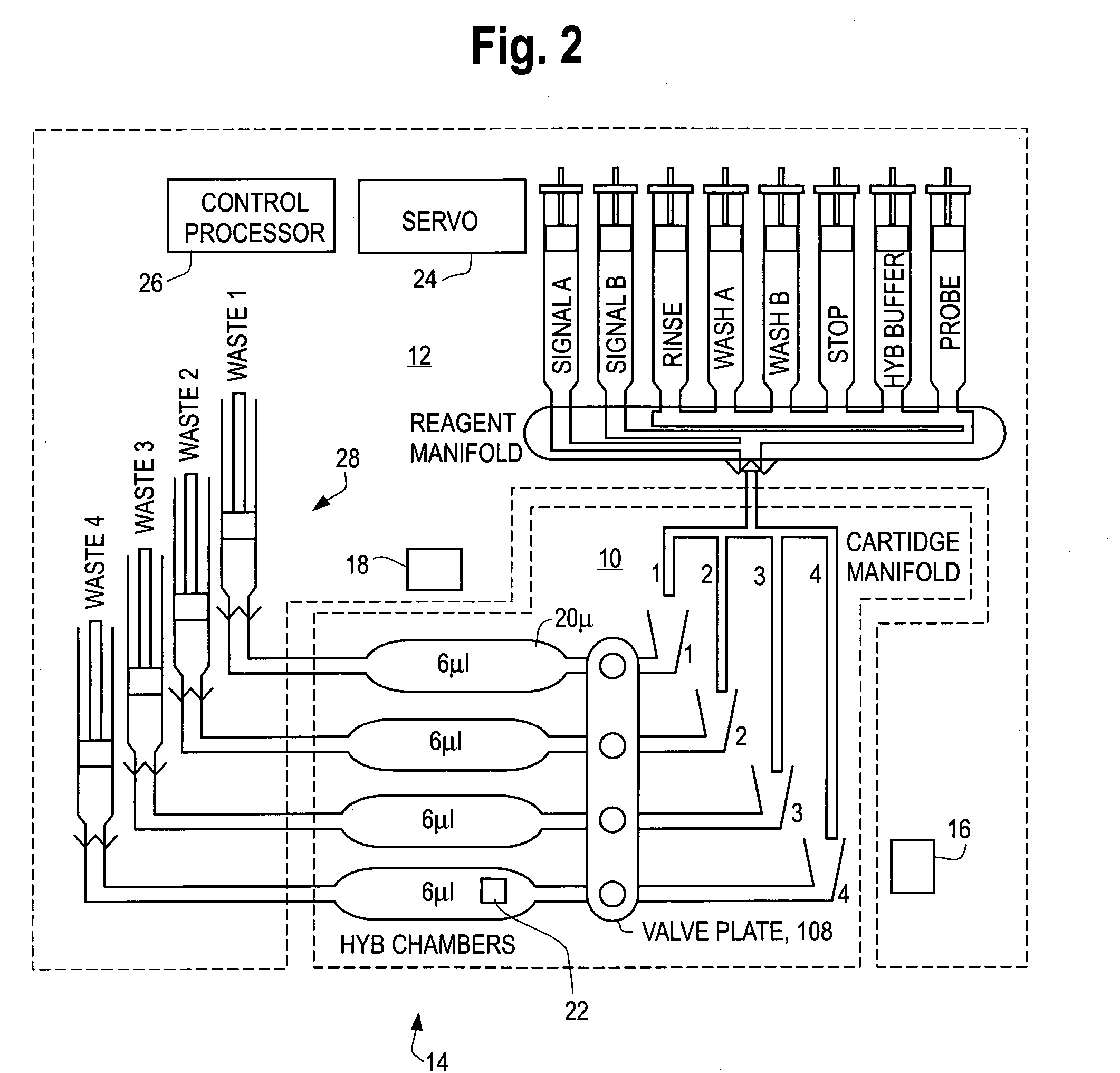
Disposable sample processing module for detecting nucleic acids
ActiveUS20050170493A1Bioreactor/fermenter combinationsBiological substance pretreatmentsA-DNABiology
A disposable sample processing module is provided for processing DNA or RNA samples. The module includes a hybridization chamber adapted to receive an oligonucleotide covalently bonded to an internal surface of the hybridization chamber. The module also include a sample well adapted to hold a DNA or RNA sample, said sample well being coupled to the hybridization chamber, a moveable valve plate disposed between the sample well and hybridization chamber, said moveable valve plate having a first position that allows transfer of the DNA or RNA sample from the sample well to the hybridization chamber and a second position that blocks transfer to the hybridization chamber and a manifold adapted to exchange fluids with the hybridization chamber to hybridize the DNA or RNA sample with the oligonucleotide, to wash the hybridized sample and to amplify the hybridized sample.
Owner:NANOSPHERE INC
Cell-based microarrays, and methods for their preparation and use
InactiveUS20060292559A1Simple methodBioreactor/fermenter combinationsBiological substance pretreatmentsImproved methodCell based
The present invention is in the field of chemistry and biotechnology. The present invention relates to cell-based microarrays, improved methods for forming such arrays, and methods for using such arrays in diagnostics, therapeutics and research. The invention particularly concerns microarrays in which ligands of a target cells are immobilized to the array support via ligand-binding molecules bound to an oligonucleotide that is hybridized to a support-immobilized oligonucleotide.
Owner:BECKMAN COULTER INC
Detection of target nucleic acid sequences by PTO cleavage and extension assay
ActiveUS8809239B2Efficient detectionMicrobiological testing/measurementLibrary creationNucleic acid sequencingBiology
The present invention relates to the detection of a target nucleic acid sequence by a PTOCE (PTO Cleavage and Extension) assay. The present invention detects a target nucleic acid sequence in which the PTO (Probing and Tagging Oligonucleotide) hybridized with the target nucleic acid sequence is cleaved to release a fragment and the fragment is hybridized with the CTO (Capturing and Templating Oligonucleotide) to form an extended duplex, followed by detecting the presence of the extended duplex. The extended duplex provides signals (generation, increase, extinguishment or decrease of signals) from labels indicating the presence of the extended duplex and has adjustable Tm value, which are well adoptable for detection of the presence of the target nucleic acid sequence.
Owner:SEEGENE INC
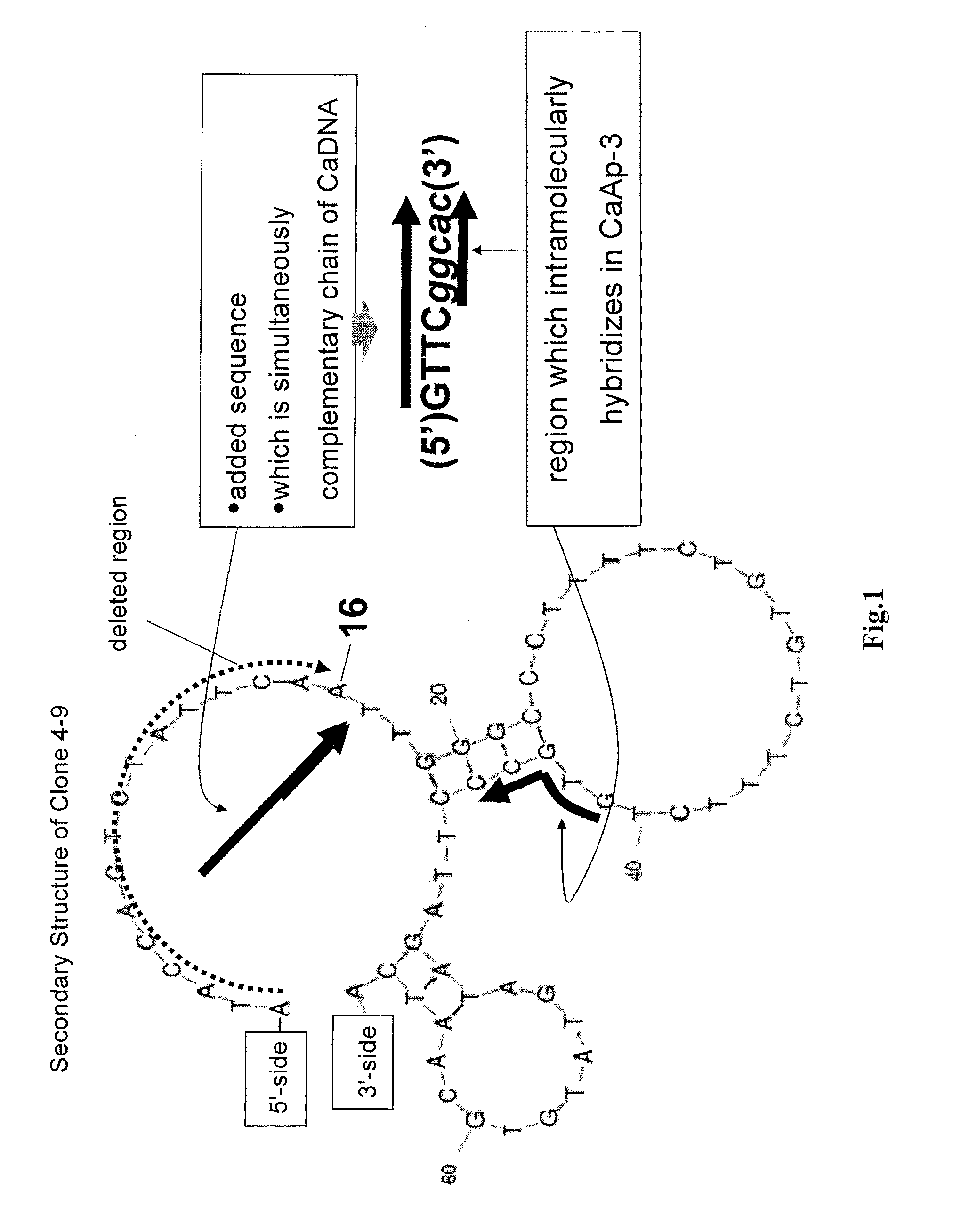
High throughput discovery of new genes from complex mixtures of environmental microbes
ActiveUS20150191720A1Raise the potentialNucleotide librariesMicrobiological testing/measurementNucleotideDecoy
Compositions and methods for isolating new variants of known gene sequences are provided. The methods find use in identifying variants, particularly homologs, in complex mixtures. Compositions comprise hybridization baits that hybridize to gene families of interest, particularly agricultural interest, in order to selectively enrich the polynucleotides of interest from complex mixtures. Bait sequences may be specific for a number of genes from distinct gene families of interest and may be designed to cover each gene of interest by at least 2-fold. Thus methods disclosed herein are drawn to an oligonucleotide hybridization gene capture approach for identification of new genes of interest from environmental samples. This approach bypasses the need for labor-intensive microbial strain isolation, permits simultaneous discovery of genes from multiple gene families of interest, and increases the potential to discover genes from low-abundance and unculturable organisms present in complex mixtures of environmental microbes.
Owner:AGBIOME INC
Bimolecular Constructs
InactiveUS20100204461A1Sugar derivativesMicrobiological testing/measurementCombinatorial chemistryFluorophore
An immobilized bimolecular construct comprises a solid support, a first oligonucleotide and a second oligonucleotide. The first oligonucleotide is labeled at one end with a fluorophore or quencher and attached at the other end to a solid support. The second oligonucleotide is labeled at one end with a fluorophore or quencher and hybridized at the other end to the first oligonucleotide. Hybridization of the second oligonucleotide with the first oligonucleotide brings the labeled end of the second oligonucleotide in close proximity or physical contact with the labeled end of the first oligonucleotide. In one embodiment the second oligonucleotide is also attached to the solid support in proximity to the first oligonucleotide. In this embodiment, the second oligonucleotide may be first attached to the solid support and then hybridized to the first oligonucleotide or, conversely, first hybridized to the first oligonucleotide and then attached to the solid support.
Owner:UNIV OF MEDICINE & DENTISTRY OF NEW JERSEY +1
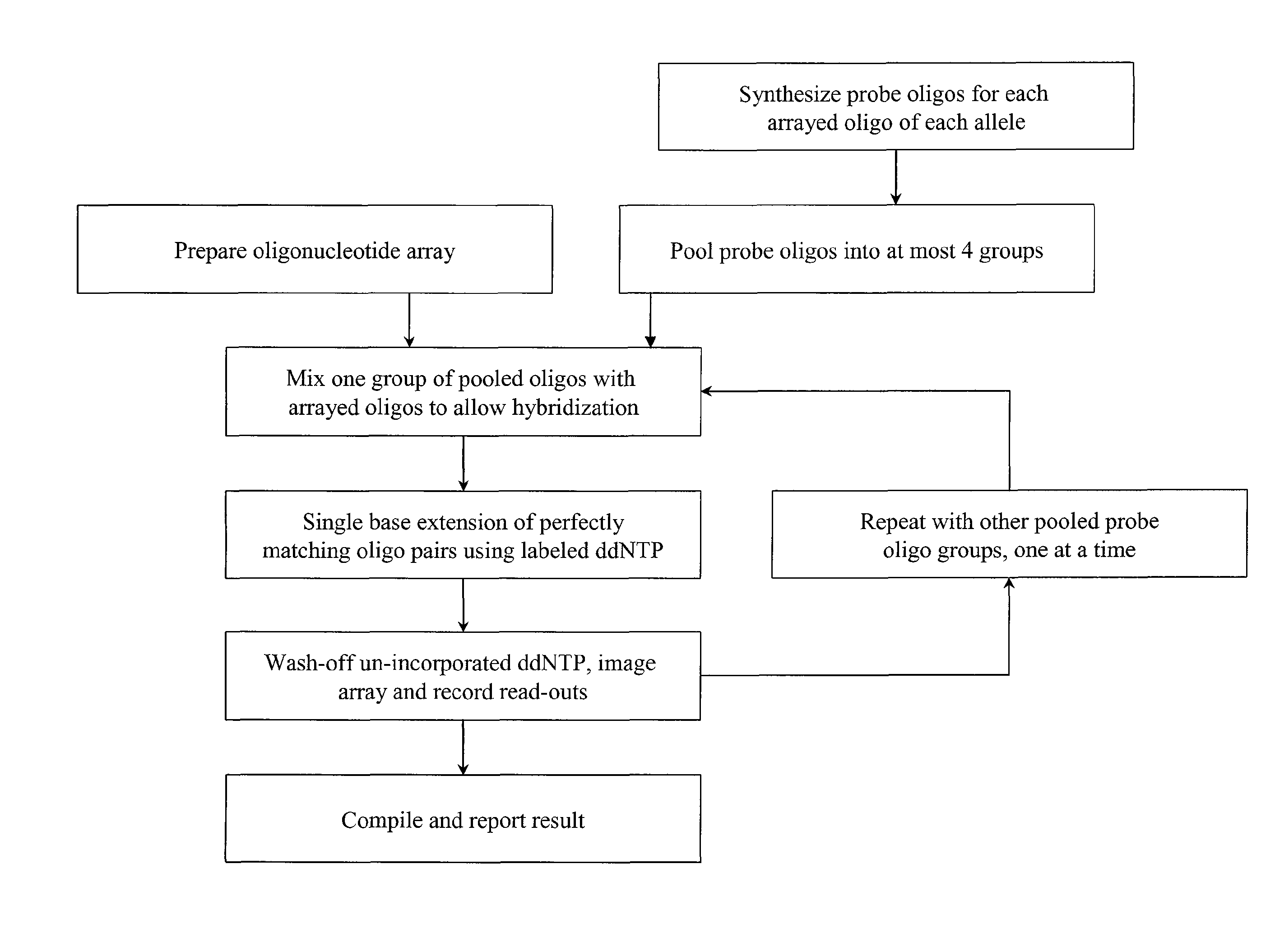
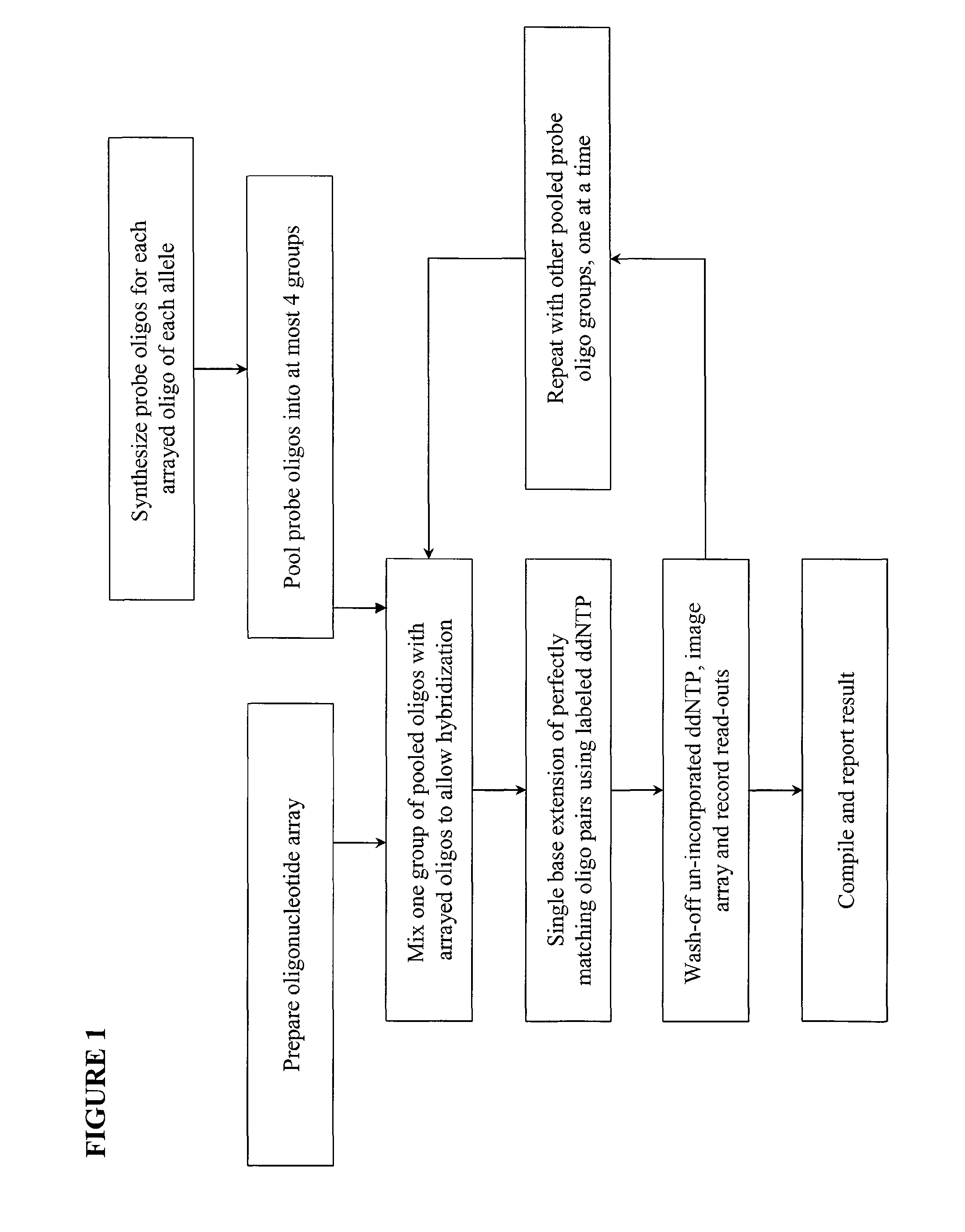
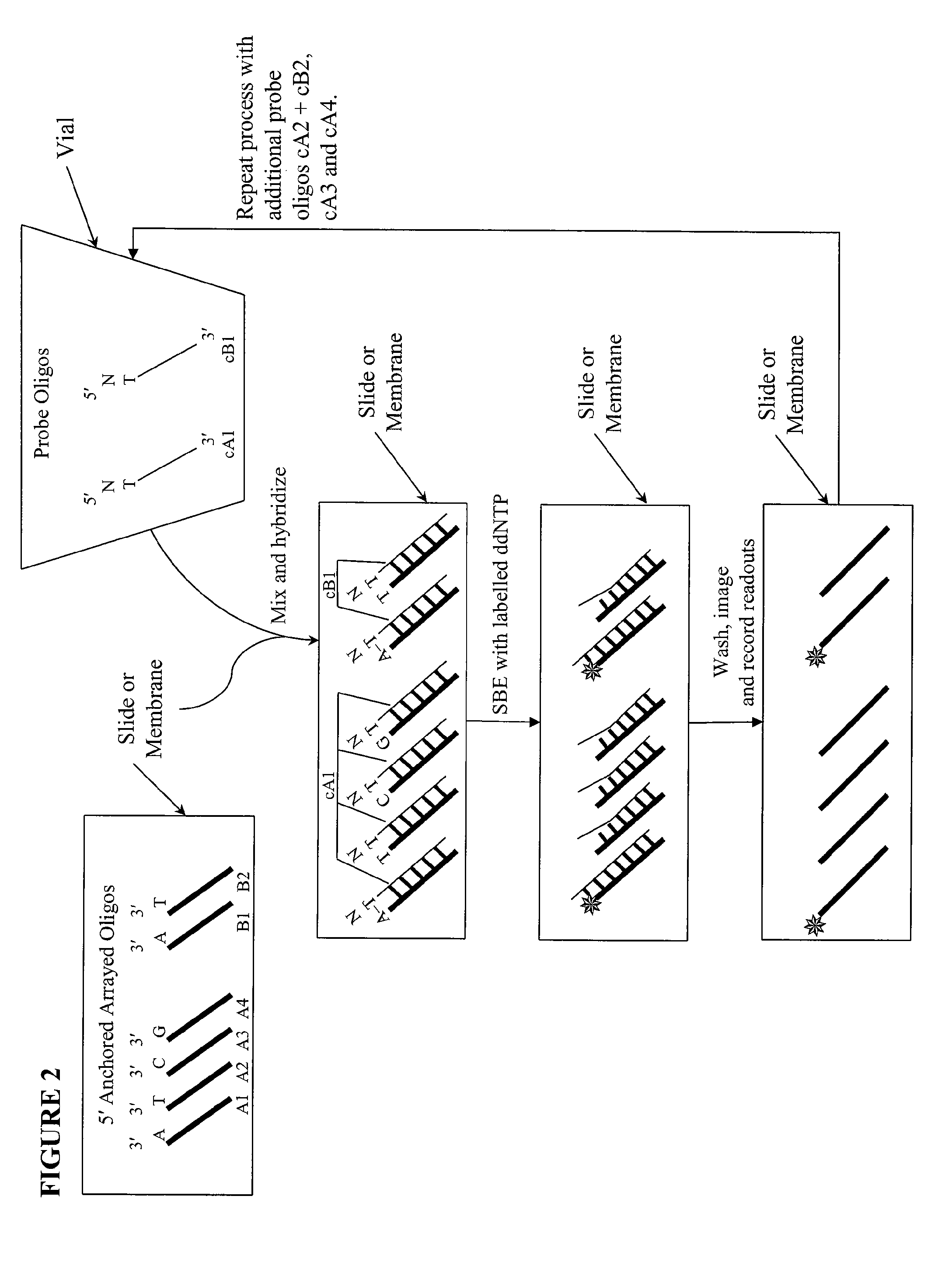
Quality control methods for arrayed oligonucleotides
InactiveUS8507197B2Quick and accurate verificationBioreactor/fermenter combinationsBiological substance pretreatmentsDideoxynucleotide TriphosphatesQuality control
We disclose quality controls methods that allow quick and accurate verification of a test oligonucleotide deposited on a solid support. It is especially useful for the verification of oligonucleotides representing alleles of a multi-allelic locus. It employs single base extension, with labeled dideoxynucleotides, to locate and verify the identity of test oligonucleotides. This approach involves synthesizing a complement probe oligonucleotide for each oligonucleotide being tested. Probe oligonucleotides are optionally grouped. They are then hybridized to test oligonucleotides, and the hybridized pair is subject to single base extension and detection. It requires the presence of one unique base, either in the last two bases at the free hanging end of the test oligonucleotide—as opposed to the end anchored to the solid support surface, or in the last two bases at one end of the probe oligonucleotide.
Owner:GE HEALTHCARE BIO SCI CORP
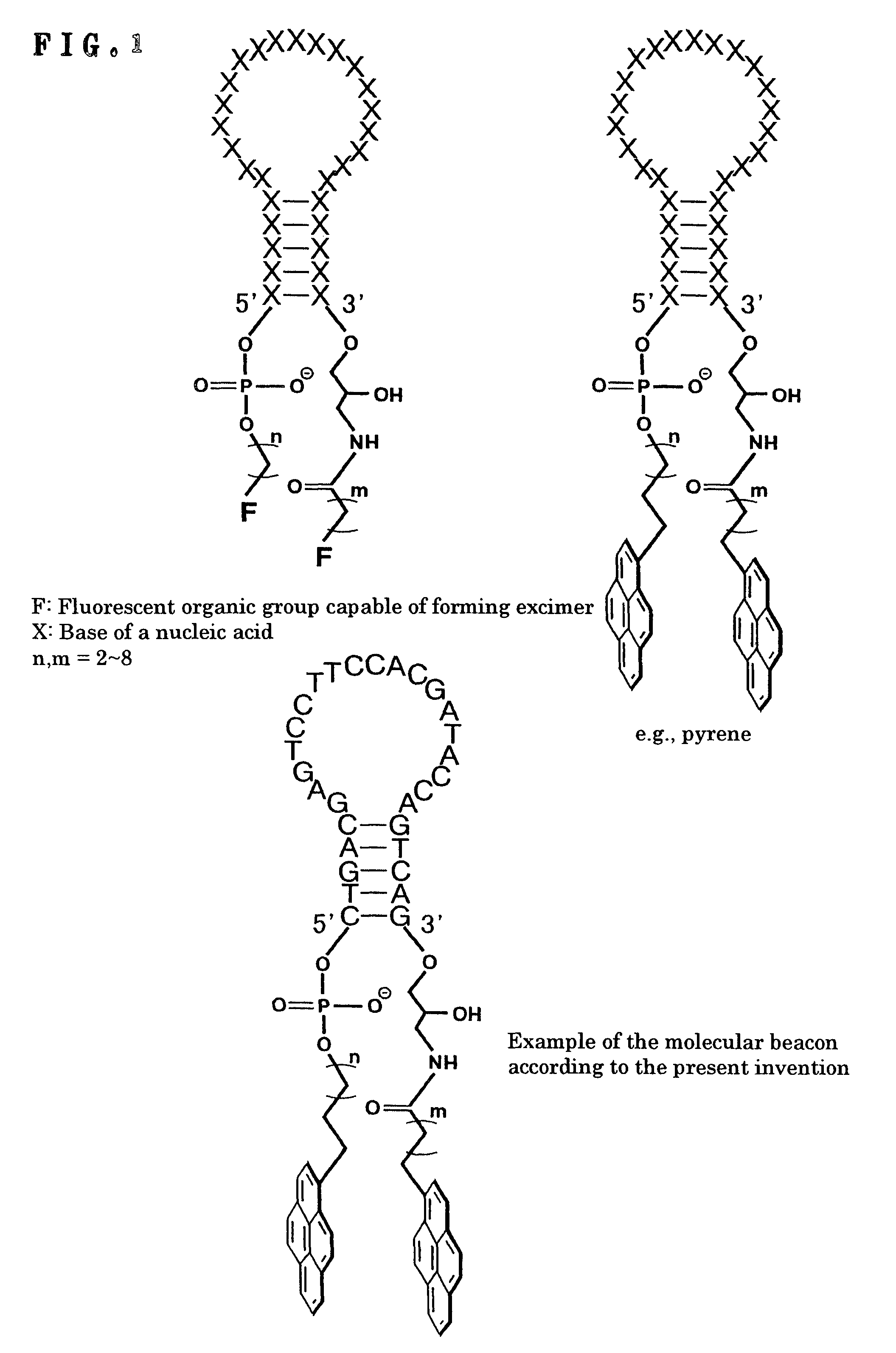
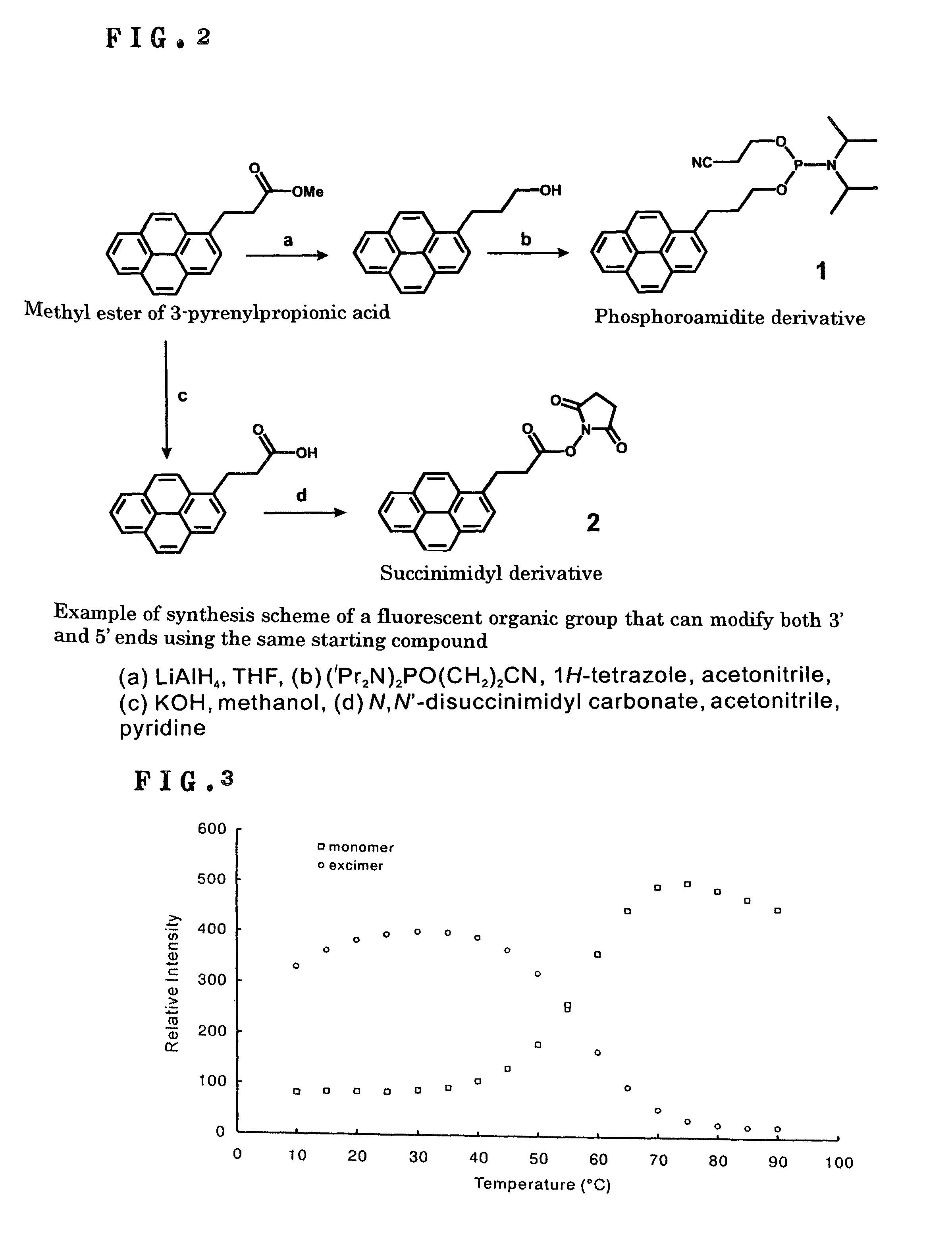
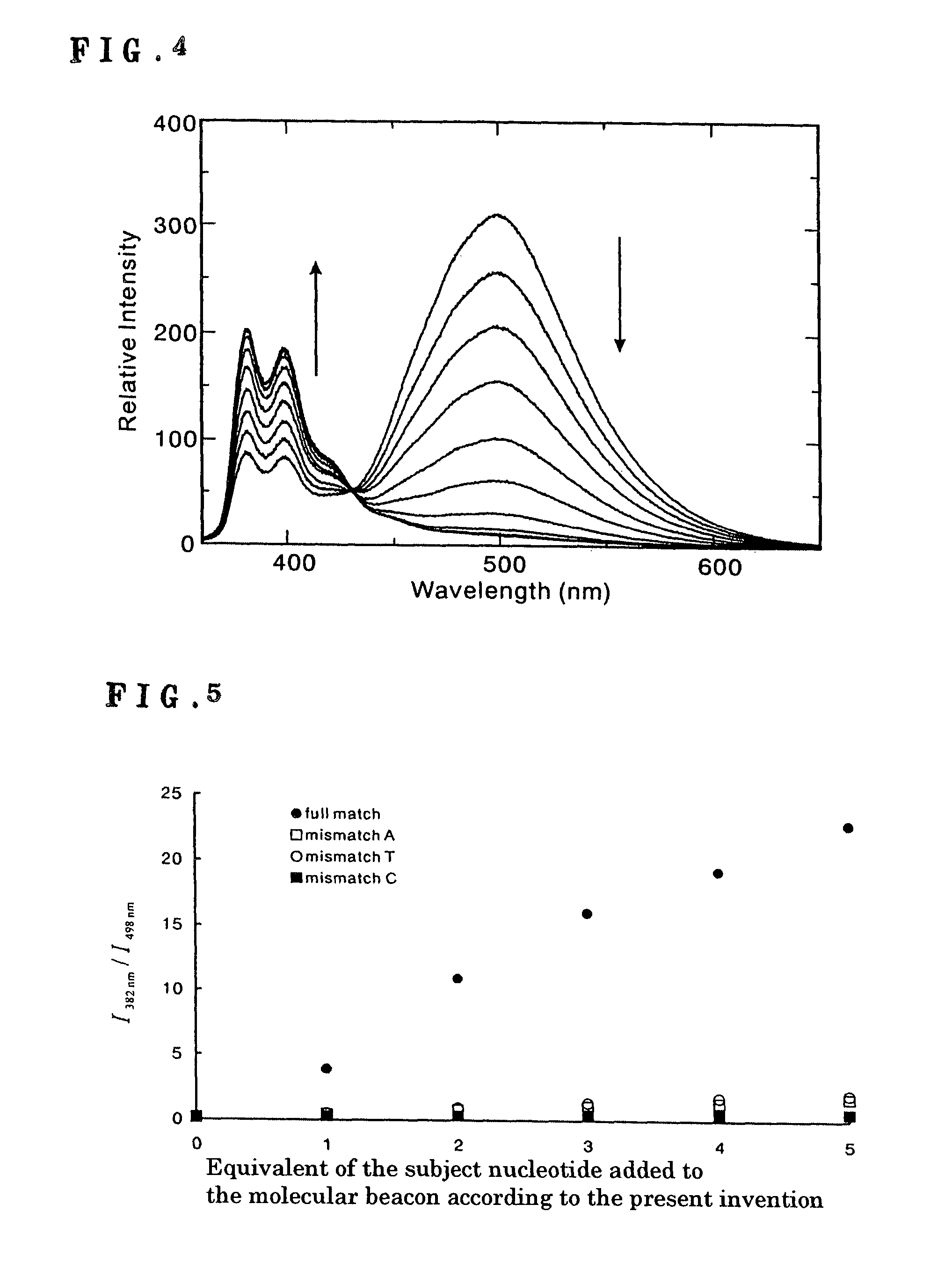
DNA detection method using molecular beacon with the use of monomer emission/excimer emission switching of fluorescent molecule
InactiveUS8105772B2Improve accuracyHigh sensitivitySugar derivativesMicrobiological testing/measurementDna testOrganic group
It is intended to provide a means and a method whereby a molecule for DNA detection at an elevated accuracy can be provided, different form the existing DNA detection system with the use of a molecular beacon. A molecular beacon wherein fluorescent organic groups capable of forming an excimer are bonded to the 3′ and 5′ ends of a single-stranded oligonucleotide to be hybridizable with a subject oligonucleotide and the switching of monomer emission / excimer emission is utilized; and a method of detecting SNP by using this molecular beacon.
Owner:JAPAN SCI & TECH CORP
Nucleic acid-based authentication codes
InactiveUS20150111780A1High sensitivityDetect changeMicrobiological testing/measurementLibrary screeningParticulatesFluorescence
This invention relates to a nucleic-acid based product authentication by determining authentication codes comprising target nucleic acids using oligonucleotide probes immobilized on particulate and non-particulate substrates. The presence of the authentication code is determined using detection methods, such as flow cytometric methods, capable of particle discrimination based on the light scattering or fluorescence properties of the particle, or by spatial resolution of oligonucleotides immobilized at specific loci on a substrate. Target-correlated fluorescence signal, originating from a target nucleic acid hybridized to substrate-immobilized oligonucleotide is determined as an indicator of the presence of the authentication code.
Owner:AUTHENTIFORM TECH
Oligonucleotide microarray technique for detecting pathogen contamination in seawater
InactiveCN101580877AStrong specificityThe detection operation process is shortNucleotide librariesMicrobiological testing/measurementDigoxinEnrichment culture
The invention relates to an oligonucleotide microarray technique for detecting pathogen contamination in seawater, belonging to the field of seawater contamination monitoring. The technique comprises the main technical schemes that a 16S-23S rRNA gene transcription interval sequence is used as a detection target and is amplified by a one-step polymerase chain reaction, a digoxin mark is obtained simultaneously, and then oligonucleotide hybridization is carried out; and the obtained monitoring result is interpreted in a manner that an enzyme-labeled antibody catalyzes the substrate colour development. Compared with the traditional product for detecting seawater contamination, the invention utilizes microarray detection to obtain the distribution situation of large numbers of pathogens and contamination index bacteria, and has the advantage of high flux; the invention can directly utilize seawater as a sample and truly obtain the contamination situation information of target bacteria under a condition of keeping the natural proportion of the flora number in the seawater; however, most existing detection techniques need the step of enrichment culture, destroy the original proportion of a flora composition, have lower reliability of the result and have longer detection procedure; and the oligonucleotide microarray detection operation has short procedure and is comparatively sensitive and fast.
Owner:NANKAI UNIV
Methods for isolating long fragment RNA from fixed samples
InactiveUS20090092979A1Sugar derivativesMicrobiological testing/measurementFormalin fixed paraffin embeddedTissue specimen
The present invention relates to methods for the extraction of long fragment RNA from fixed tissue specimens. In particular, the present invention relates to methods for the extraction of RNA from formalin-fixed paraffin-embedded tissue specimens for use in biologic applications, including assays based on oligonucleotide hybridization.
Owner:RESPONSE GENETICS
Nucleic acid-based linkers for detecting and measuring interactions
ActiveUS9914958B2Eliminate errant dataReliable linkMicrobiological testing/measurementBiological testingAptamerNucleotide
The invention provides compositions comprising nucleic acid complexes for use in monitoring binding interactions and in measuring association and / or dissociation kinetics with or without force, detecting analytes, screening aptamers, and encoding / encrypting information. In some instances, the nucleic acid complexes are double-stranded nicked nucleic acids comprising a scaffold nucleic acid hybridized to one or more oligonucleotides. In some instances, a first and / or a second oligonucleotide are linked to moieties that are known to interact with each other or which are suspected of interacting with each other.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +1
Method for analysing the interaction of nucleotide sequences in a three-dimensional DNA structure
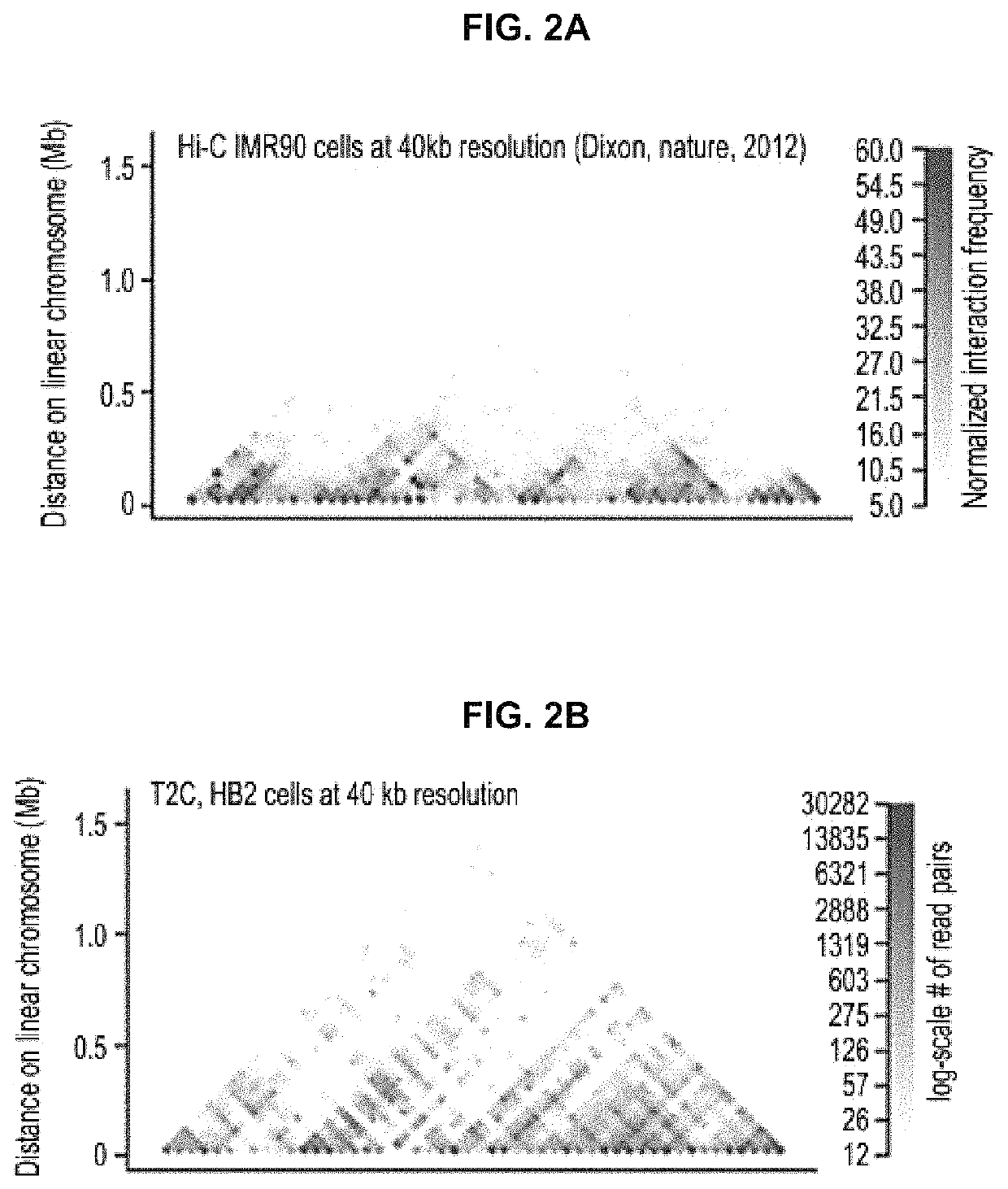
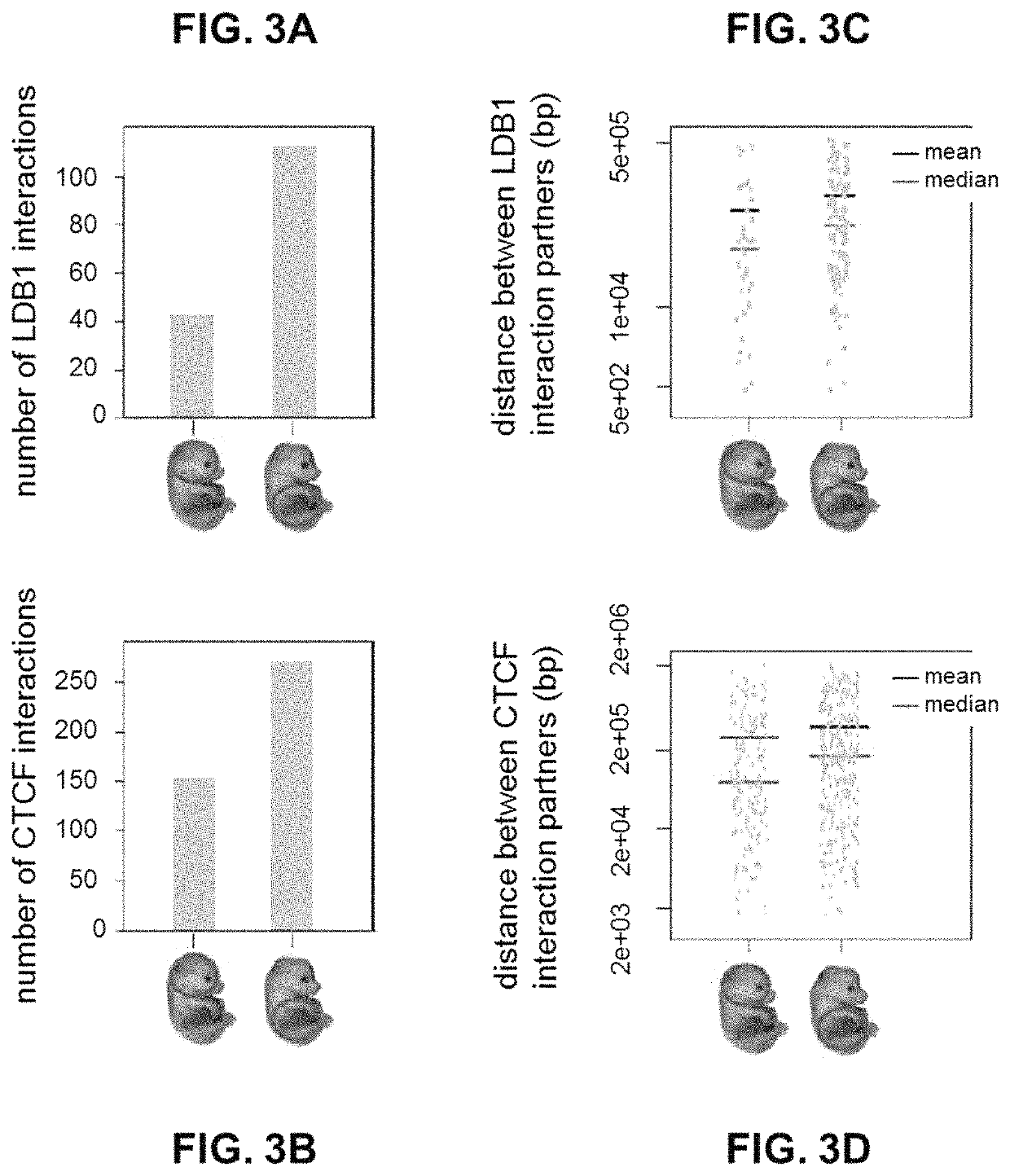
ActiveUS10934575B2Increase the number ofHigh resolutionMicrobiological testing/measurementRestriction enzyme digestionNucleotide
The present invention provides a method for analysing the interaction of one or more nucleotide sequence(s) from one or more region(s) of interest with other nucleotides sequences in a three-dimensional DNA structure, comprising the steps of: (a) providing a sample of crosslinked DNA; (b) digesting the cross-linked DNA with a first restriction enzyme; (c) ligating the cross-linked nucleotide sequences; (d) reversing the cross-linking; e) fragmenting the ligation and ligated molecules from (d); (f) hybridising the fragments from (e) to one or more oligonucleotides representing the sequences which are adjacent to the cleavage site of the first restriction enzyme in order to enrich for the ends of the nucleotide sequences that have been ligated to another nucleotide sequence in step (c); and (g) analysing the nucleotide sequence of the enriched fragments in order to identify the nucleotide sequences involved in interaction(s).
Owner:ERASMUS UNIV MEDICAL CENT ROTTERDAM ERASMUS MC
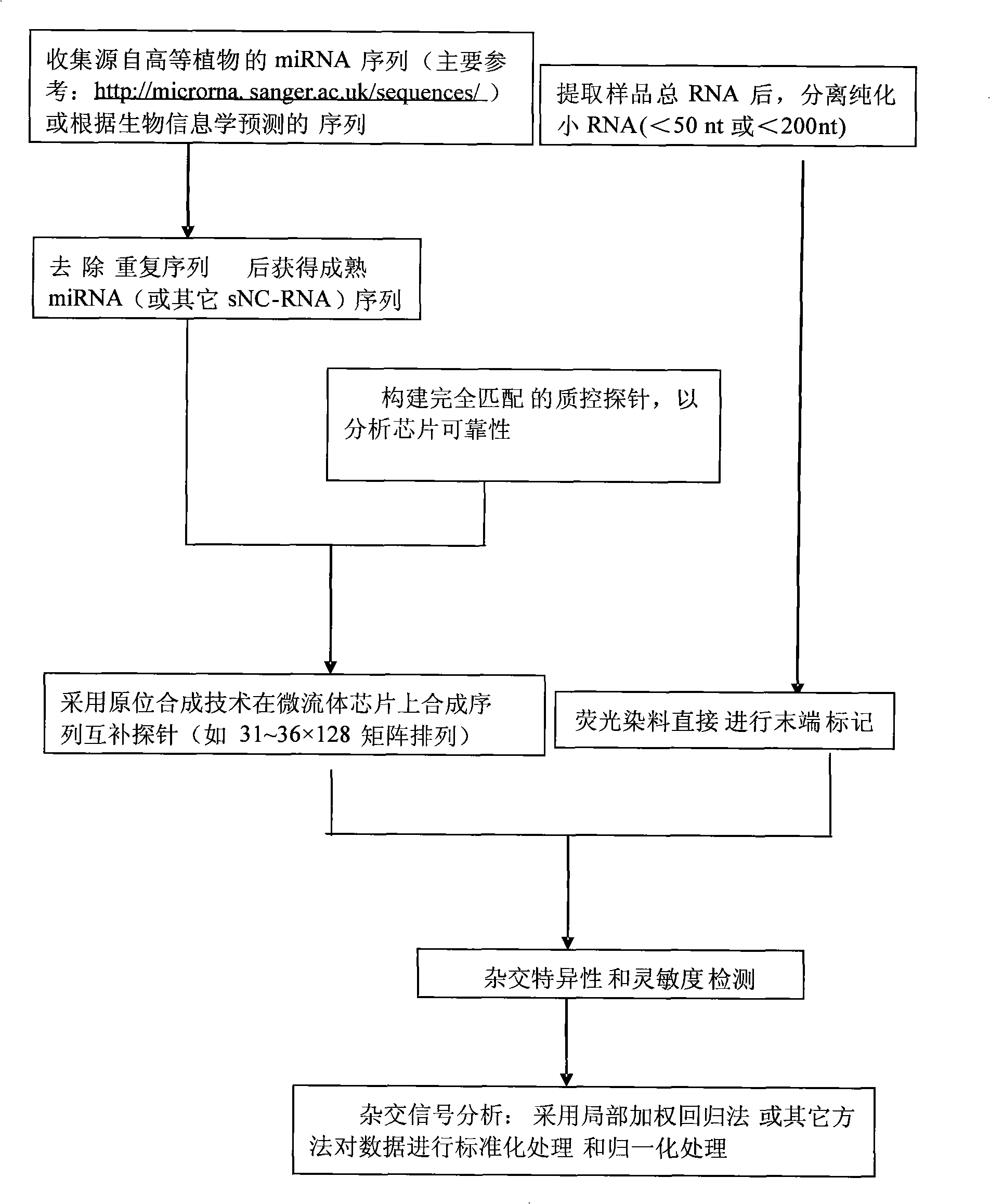
Detection process for synthesizing microfluid chip by in-situ method
InactiveCN101285100AHigh sensitivityImprove throughputMicrobiological testing/measurementOligonucleotide chipSense strand
The invention discloses a detection method synthesizing microfluidic chips in situ, which comprises the following steps that: 1) a sequence of non-coding small RNAs reported on the prior model plant is utilized, sequence information of miRNA is preferably selected, and complementary oligonucleotide hybridization probes are designed according to the sequence of a sense strand or an antisense strand of miRNA; 2) an oligonucleotide chip is prepared by an in-site synthetic method; 3) small RNAs smaller than a 50 oligonucleotide group are extracted from a plant to be detected, then are hybridized with the probes in the chip after making end labeling, and the result of chip hybridization is obtained to perform qualitative and quantitative analysis. The detection method can be used for the validation of miRNA (and other sNC-RNAs) obtained through the forecast by a bioinformatics method, can also be used for the detection and identification of miRNA (and other sNC-RNAs) in new plant species, and can also be used for the determination of miRNA (and other sNC-RNAs) variation relation under different treatment conditions (eg. under the condition of virus infection).
Owner:ZHEJIANG SCI-TECH UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com