Detection of nucleic acids
A target nucleic acid and detector technology, which is applied in the determination/inspection of microorganisms, measurement devices, preparation of samples for testing, etc., can solve problems such as inability to detect target nucleic acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
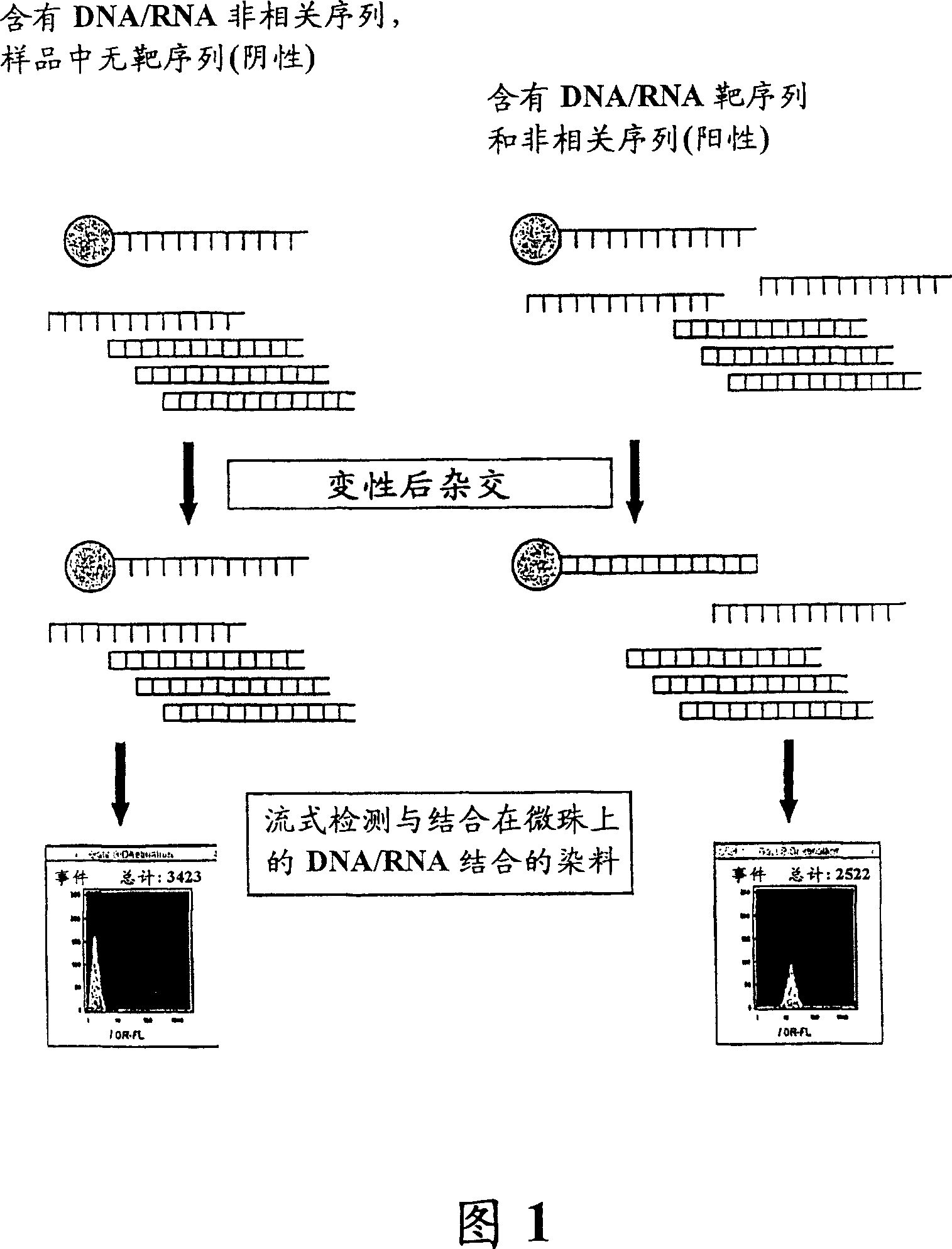
[0179] Detection of target DNA hybridized to particle-immobilized oligonucleotides: DNA-binding fluorophores
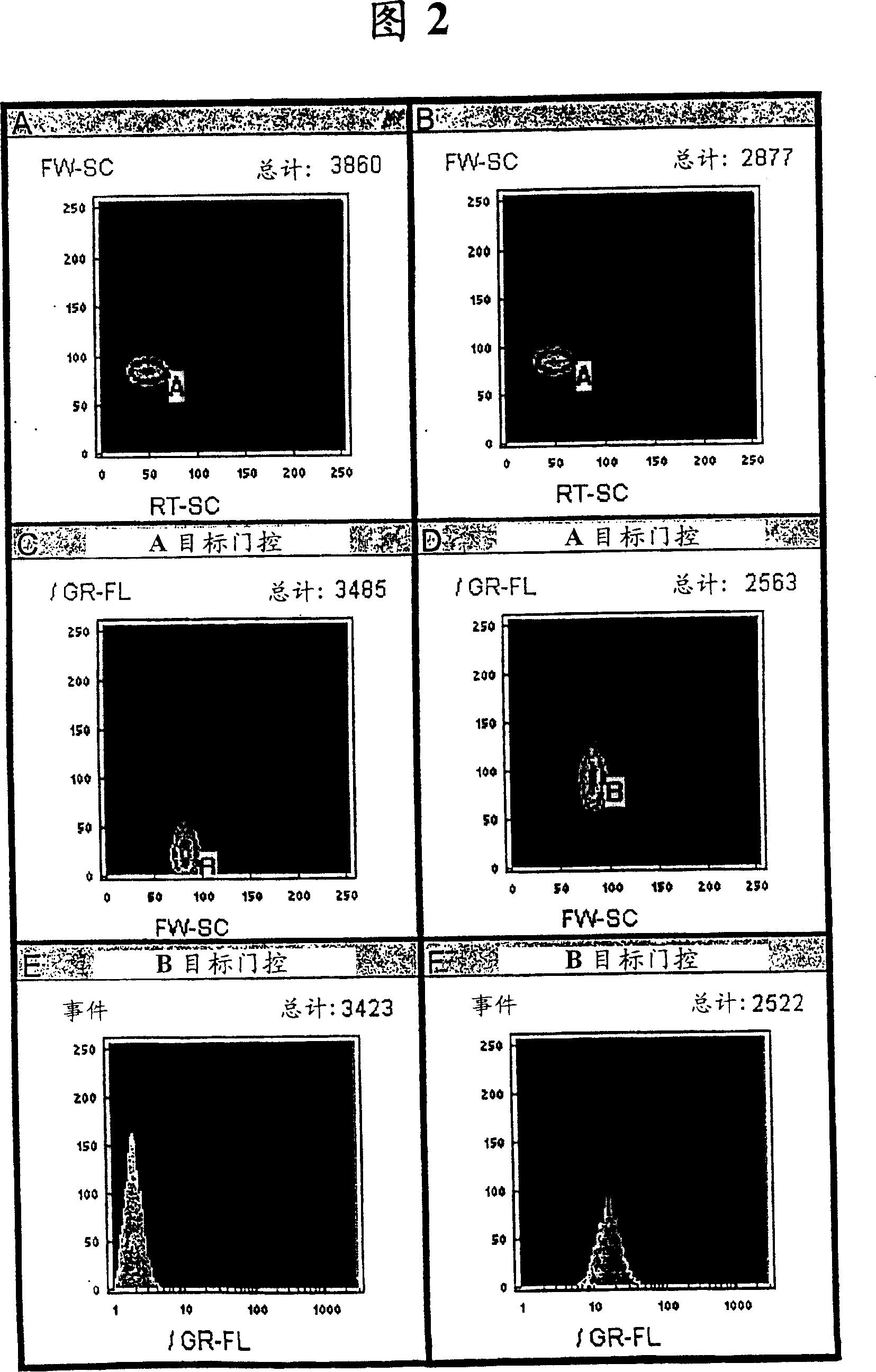
[0180]Hybridized target DNA is detected with a fluorophore that binds to dsDNA. The target DNA (SEQ ID NO: 2) in this example was hybridized to the bead-immobilized probe (SEQ ID NO: 1) in the presence of excess calf thymus DNA. Fluorophores are mixed with the hybridized target-bead suspension and analyzed by flow cytometry. The results using thiazole orange as the dsDNA binding fluorophore are shown in Figure 2A-F. Front azimuthal scatter (FW-SC) and right angle scatter (RT-SC) patterns of beads incubated with 1 μg of calf thymus DNA in the absence (Fig. 2A) and presence (Fig. 2B) of target DNA are shown. Light scatter gating of the beads allows analysis of particle sorting within a narrow range of FW-SC x RT-SC values by means of image analysis software algorithms, thus eliminating particles outside that size range. Using thiazole orange as the dsDNA binding fl...
Embodiment 2
[0185] nuclease protection
[0186] Hybridization of the biotinylated particle-immobilized oligonucleotide probe (Oligo-1A) to the target oligonucleotide (SEQ ID: 2) protects the probe from hydrolysis by single strand specific DNA endonucleases. Particle-immobilized oligonucleotides and CTDNA were incubated together in the presence and absence of 1000 femole target DNA as described in Example 1, followed by incubation with S1 nuclease. Biotin is released upon endonuclease hydrolysis of the oligonucleotide probe unless it is protected from hydrolysis by hybridization to the target DNA.
[0187] Add 8 µl of an aliquot of streptavidin-linked fluorescer (streptavidin-phycoerythrin from Molecular Probes) diluted 1:10 in TE buffer to 16 µl of nuclease-treated Treated samples were incubated at room temperature for 10 min. The bound streptavidin-linked fluorescer serves as a reporter for the intact bead-linked oligonucleotide. The mixture was analyzed by flow cytometry. The mean c...
Embodiment 3
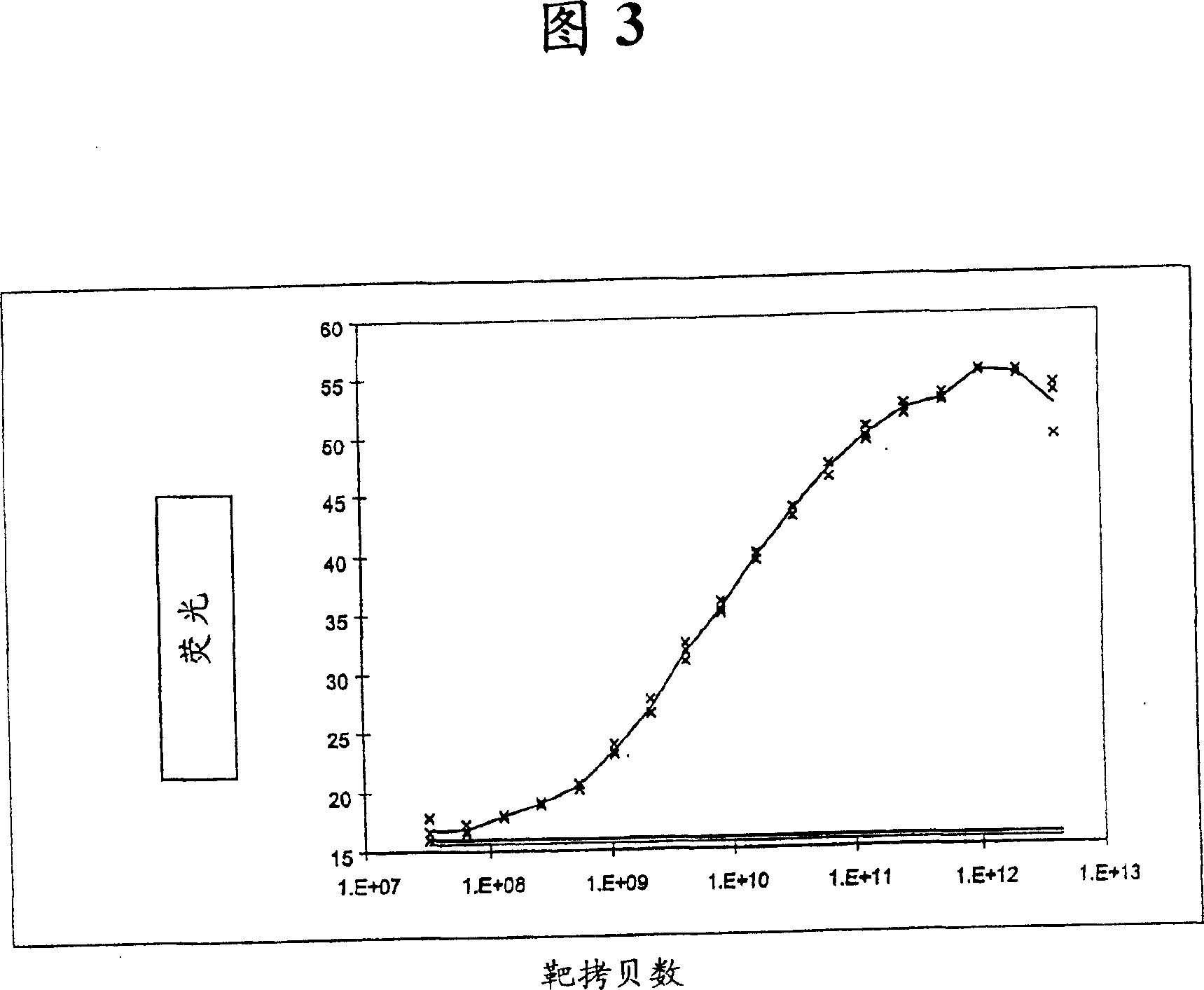
[0189] Quantification of target DNA: DNA-bound fluorophores
[0190] As shown in Figure 3, the fluorescence of the fluorophore sybr green bound to a hybrid of target DNA (SEQ ID NO: 2) and bead-immobilized oligonucleotide (SEQ ID NO: 1) varies with target copy number .
[0191] The uniqueness of the fluorescent signal associated with bead-immobilized oligonucleotides was dependent on the concentration, demonstrating that the target DNA content can be quantified over a concentration range of approximately 6 orders of magnitude. In the background of 0.8 μg of non-specific calf thymus DNA, it was clearly detected as low as the equivalent of 440 attomoles (attomoles) or about 2.5×10 8 25.7 picograms of target dsDNA were copied for the 90-mer target. Therefore the target DNA is at about 3.11 x 10 4 It is readily detectable in the presence of a fold excess of nonspecific DNA. The results also show that the method is very sensitive in the concentration range; about 1.25×10 8 to ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com