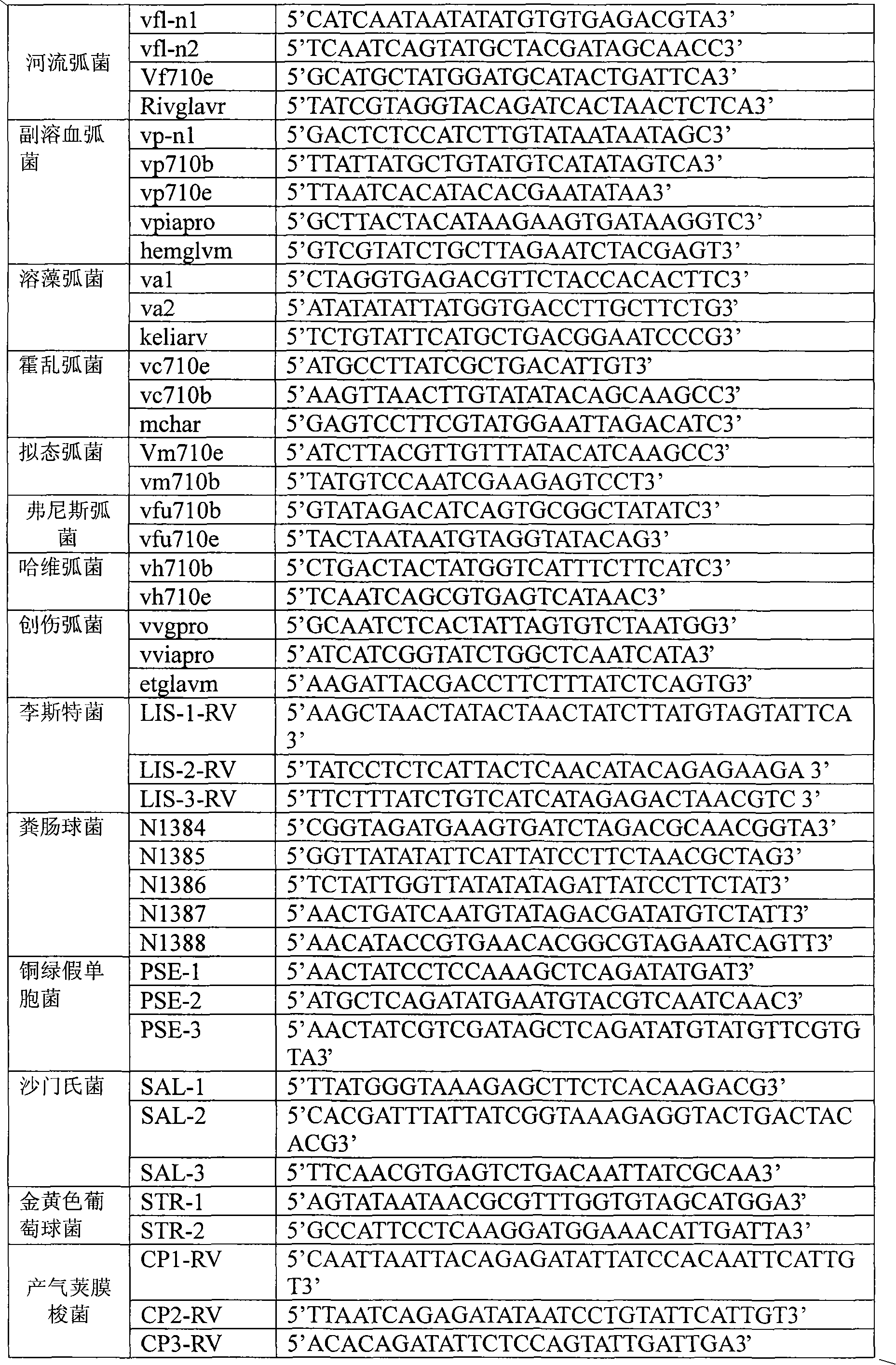
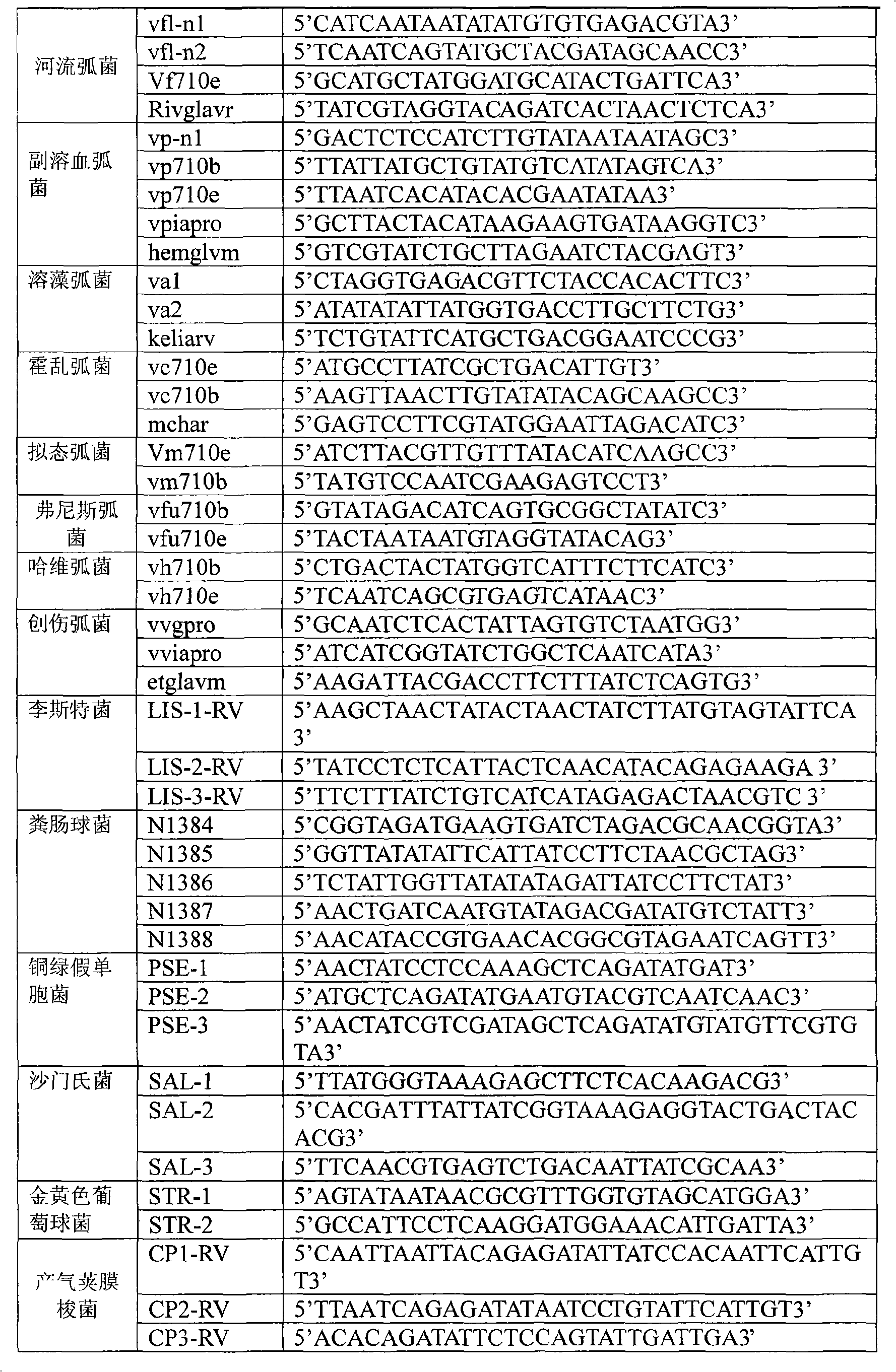
Oligonucleotide microarray technique for detecting pathogen contamination in seawater
An oligonucleotide and microarray technology, applied in the field of seawater pollution monitoring, can solve the problem that the overall pollution status of seawater pathogens cannot be effectively given.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Embodiment 1: seawater sample 1
[0072] 1 Oligonucleotide Microarray Fabrication:
[0073] 1) Preparation of probe dilution: 1× salmon sperm DNA solution (1g / L)
[0074] 2) Prepare probe solution: use probe diluent to prepare 0.1ug / μL probe solution
[0075] 3) Immobilizing the probe on the nylon membrane substrate: spot the probe solution on the 5cm×5cm nylon membrane substrate, and keep warm at 80°C for 2 hours.
[0076] 2 Seawater sample processing and DNA extraction
[0077] 1) Sample processing
[0078] ①Take 20L of surface seawater and pack it into sterilized plastic buckets.
[0079] ②Filter with a 0.15mm pore size sieve to remove suspended matter, and then use a Mini-pellicon system (Millipore, USA) to carry out pressure filtration to collect the bacteria. The filter membrane has a pore size of 0.22 μm (Milipo, USA). Every 2L of seawater filtered corresponds to a filter membrane, that is, every 2L of seawater is taken as a sample, and each sampling point c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com