Allele variant detection method, kit and composition
An allele and detection method technology, applied in the direction of recombinant DNA technology, DNA/RNA fragments, etc., can solve the problem of low detection rate, and achieve the effect of improving sensitivity and specificity, and improving amplification efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
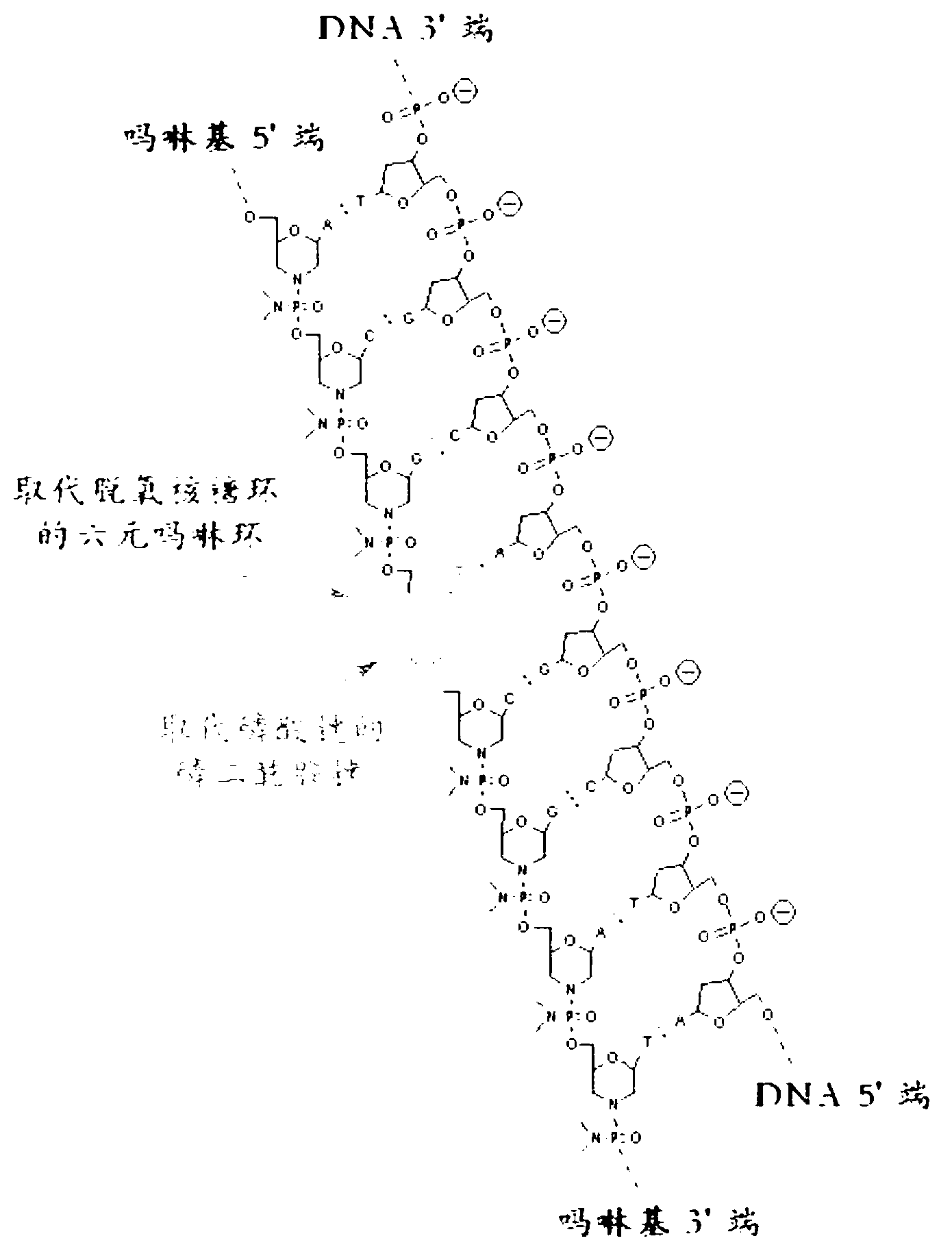
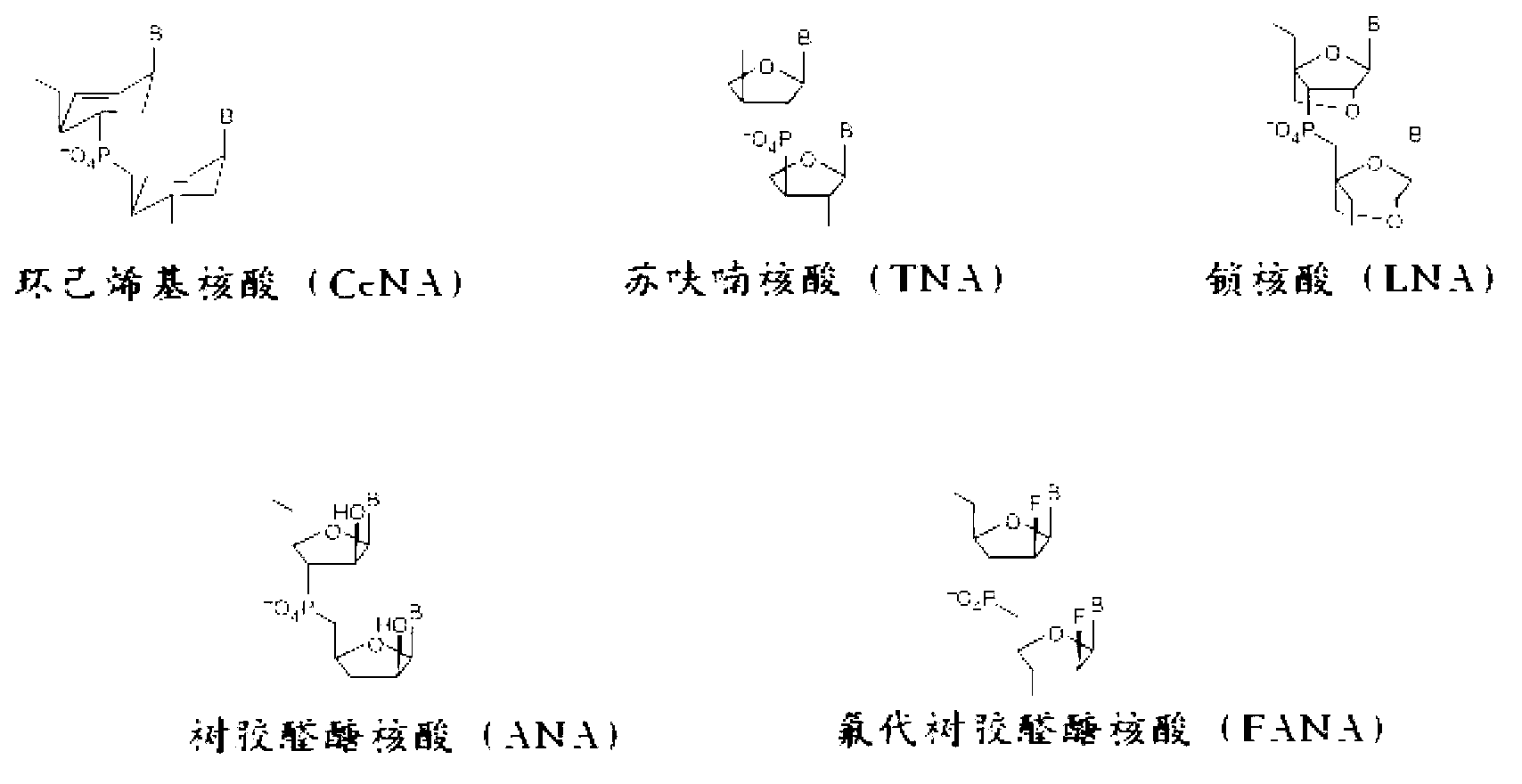
[0051] The present invention mainly relates to detection methods, kits and compositions of allelic variants. The main terms involved have the following definitions:
[0052] Allelic variant: A variant form of one of two or more genes that occurs at a particular locus on a chromosome, including single nucleotide polymorphisms (SNPs), insertions, inversions, Bits and missing.
[0053] Primer: An oligonucleotide that hybridizes to a strand of nucleotides in a target sequence.
[0054]The alleles detected in the present invention are derived from accounting samples, and the accounting samples can use any nucleic acid molecule, which can be DNA, such as genomic DNA (gDNA) or complementary DNA (cDNA), or RNA, such as messenger RNA ( mRNA), etc.
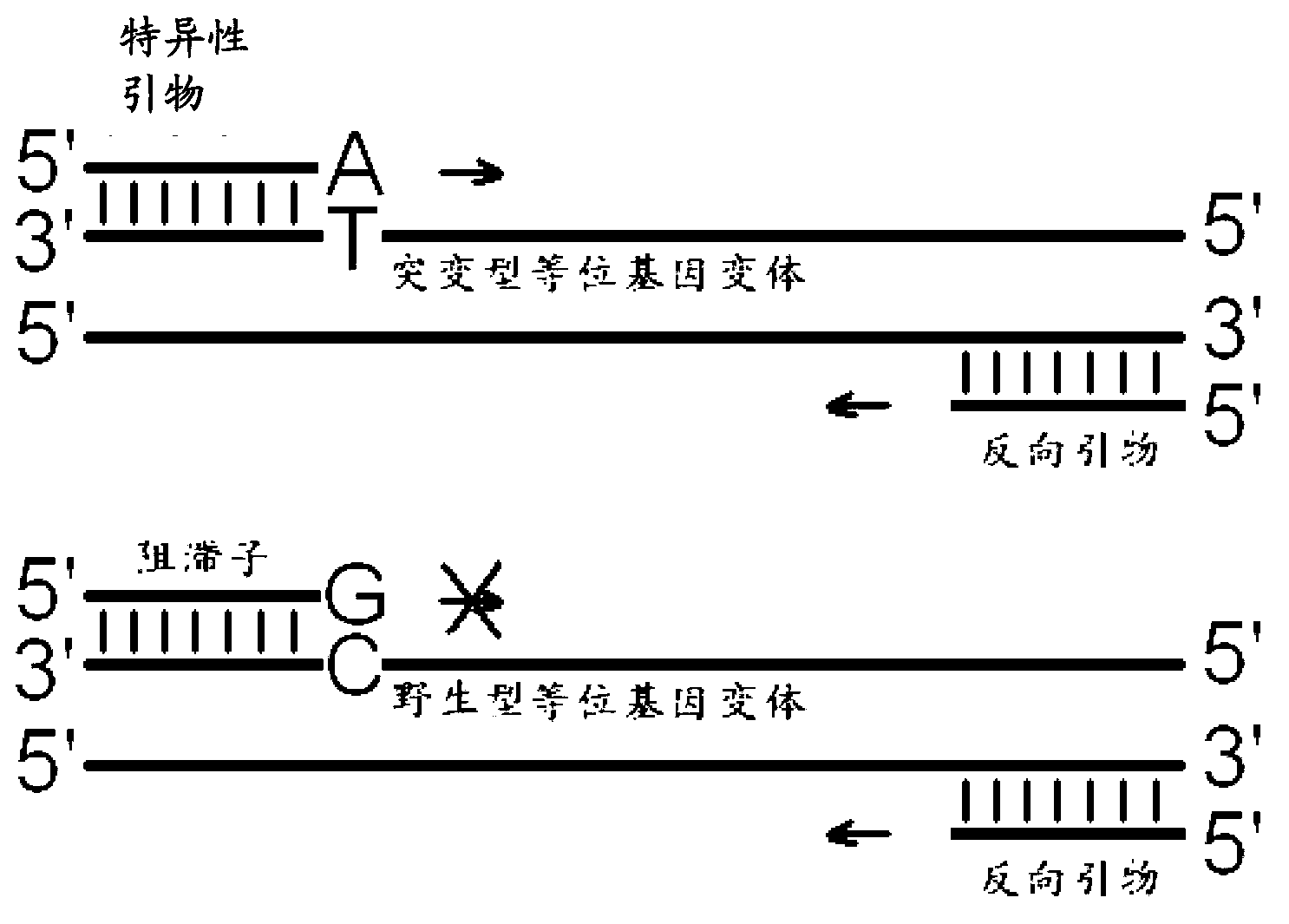
[0055] The allelic variant detection method of the present invention specifically comprises the following steps:
[0056] 1) Add mutant allele-specific primers and their reverse primers and a blocker whose 3' end cannot be extended by p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com