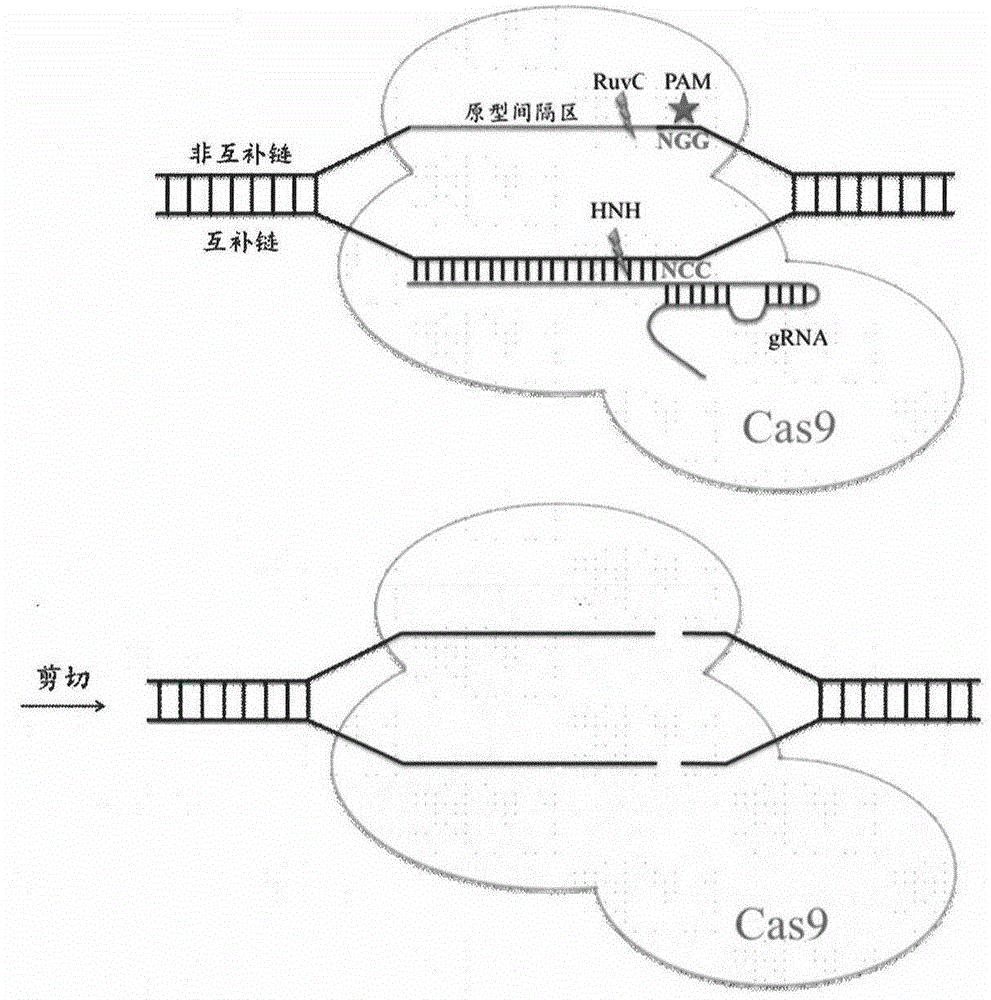
Novel mitochondrial genome editing tool
A mitochondrial genome, mitochondrial technology, applied in the field of genome engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] A new type of mitochondrial genome editing tool, the construction of the mtCRISPR / Cas9 system. 1.1 The construction of mtCas9 protein expression module:
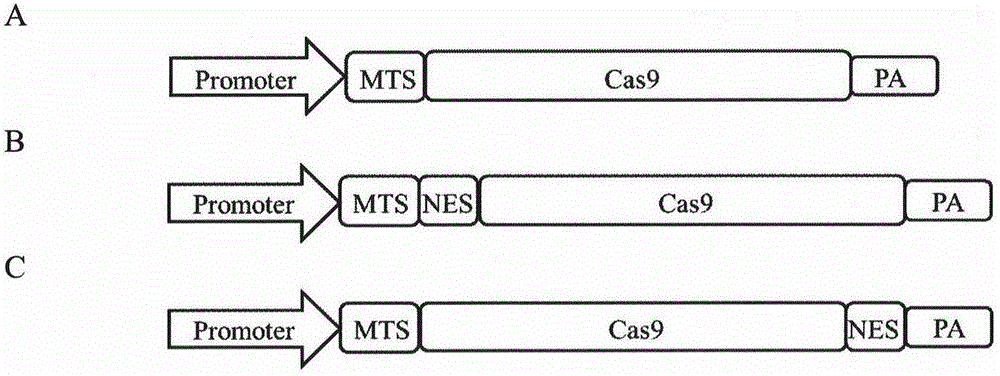
[0051] The Cas9 nuclease located in the mitochondria can be located in the mitochondria (such as image 3 Shown in A). The MTS fused to its N-terminal and NES fused to its N-terminal or C-terminal and other positions can also work together. NES can make Cas9 nuclease more distributed outside the nucleus, so that mtCas9 is better positioned in the mitochondria (Such as image 3 B and 3C).
[0052] The two mtCas9 protein expression modules constructed in this example are as follows: image 3 Shown in A and 3B. Among them, the promoter is CBh, MTS is the mitochondrial leader peptide signal of human ornithine carbamoyltransferase (Homosapiensomithinecarbamoyltransferase, OTC), NES is the strong nuclear signal in the HIV-1 rev domain, and the Cas9 coding sequence is the humanized Cas9 The coding sequence, PolyA is a polyaden...
Embodiment 2
[0091] A new type of mitochondrial genome editing tool, the mtCas9 protein co-localizes with mitochondria in the mtCRISPR / Cas9 system.
[0092] 2.1 Immunofluorescence experiment to analyze the co-localization of mtCas9 protein and mitochondria:
[0093] Specific steps are as follows:
[0094] Inter-cell operation: 1) Put the cover glass into the six-well plate, and implant HEK293 cells of appropriate density. The plasmids containing two protein expression modules were transiently transfected the next day, namely: CBh-MTS-Flag-hSpCas9-bGHpolyA and CBh-MTS-NES-Flag-hSpCas9-bGHpolyA, named MTS-Cas9, MTS-NES- Cas9; 2) After the cells are cultured for a suitable time, the cells are taken out, the cells are washed twice with PBS, and then the mitochondrial specific dye MitoTracker diluted and mixed with the medium in advance is added, and the cells are allowed to stand for 20 minutes.
[0095] Intercellular operations: 1) Cell fixation: Take out the cells, wash the cells twice with PBS, dr...
Embodiment 3
[0104] A new type of mitochondrial genome editing tool, the mt-gRNA in the mtCRISPR / Cas9 system can enter the mitochondria across the membrane.
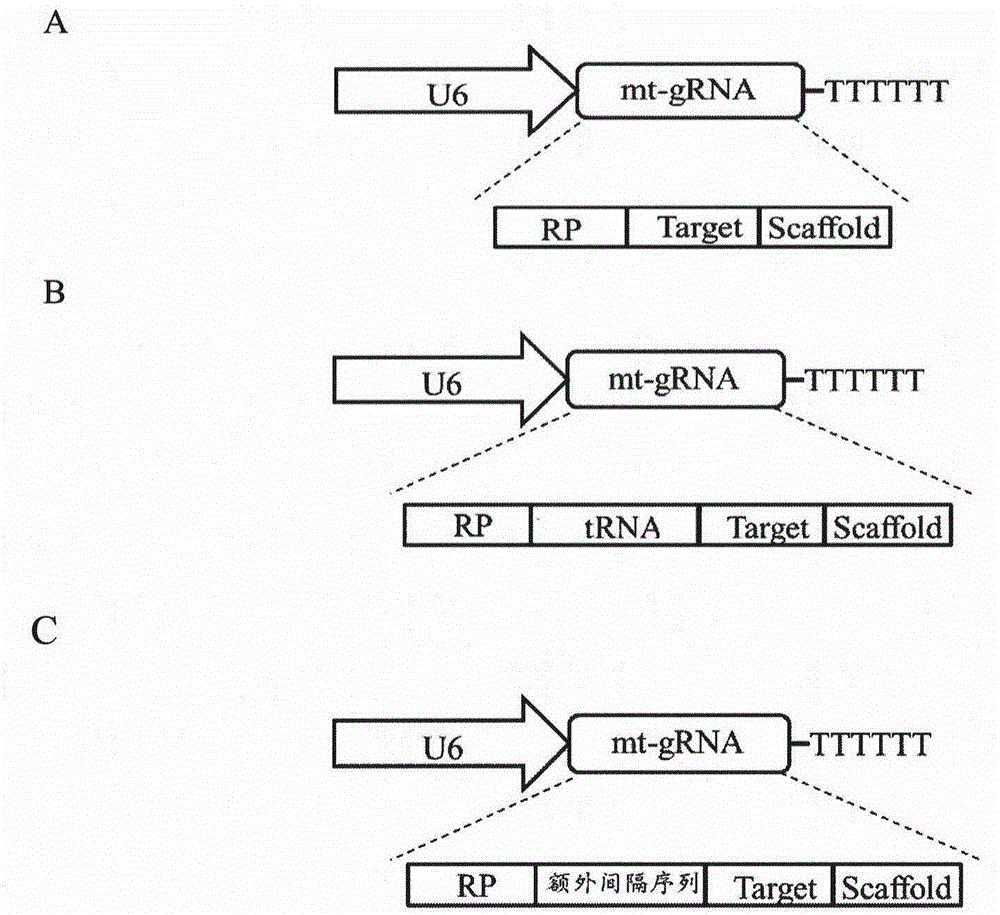
[0105] The guide RNA that enters the mitochondria in the present invention includes two types, and the specific construction method is as described in 1.2 in Example 1. The mt-gRNA of the plasmid used in this example was constructed by the second construction method with tRNA-Leul, in which the selected target sequence was a segment within the range of mitochondrial 4977bp CommonDeletion, named T1 (the position of action is as Figure 7 As shown in A, the nucleotide sequence of T1 is shown in nucleotides 7-26 in SEQ ID NO. 74), and this plasmid is named mtCRISPR / Cas9-RP-tRNA-T1.
[0106] Verification steps: 1) Transfect mtCRISPR / Cas9-RP-tRNA-T1 into HEK293 cells, harvest the cells 24h later, separate mitochondrial components, and extract total RNA (Total-RNA) and mitochondrial RNA (Mito-RNA); 2) Use reverse primers 18S-Rn, 12S-Rn, and g-R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com