Qualitative and quantitative detection method for paddy microorganisms
A technology for microorganisms and target microorganisms, applied in the biological field, can solve the problems of inaccurate qualitative and quantitative detection of microorganisms, and achieve the effects of fine detection results, simple detection process, and probability guarantee.
Inactive Publication Date: 2016-05-25
INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI +1
View PDF4 Cites 3 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0004] In order to solve the problem of inaccurate qualitative and quantitative detection of microorganisms in the prior art, the embodiment of the present invention provides a qualitative and quantitative detection method for rice microorganisms
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
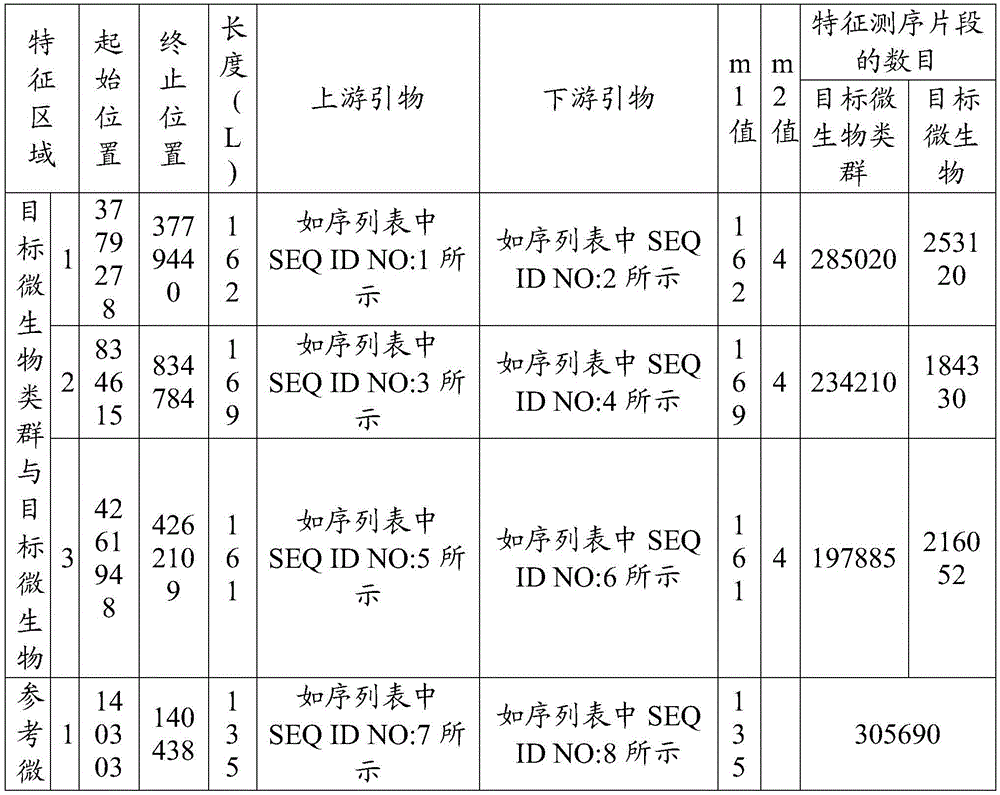
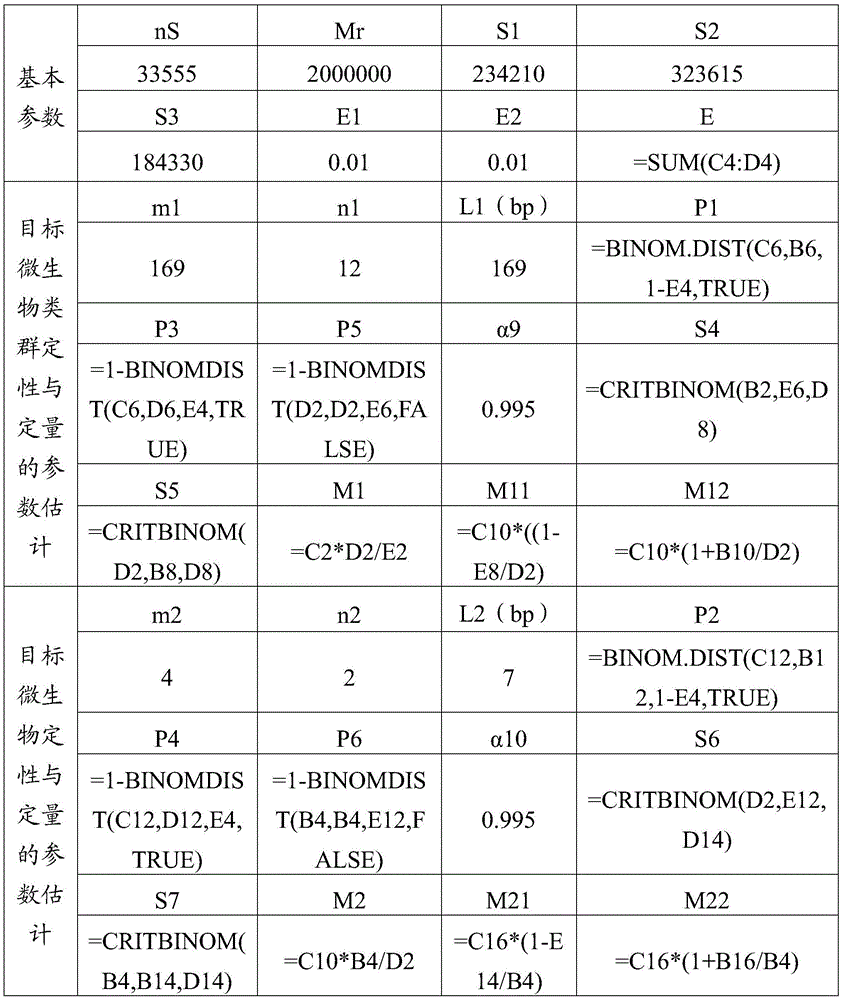
[0040] Identification of Xanthobacterium oryzae
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses a qualitative and quantitative detection method for paddy microorganisms and belongs to the technical method of biology. The method comprises the following steps: determining target microorganism groups, target microorganisms and non-target microorganisms of a to-be-measured sample and reference microorganisms non-existing in the to-be-measured sample; designing characteristic regions of the target microorganism groups and the target microorganisms; designing multi-amplification primers of the characteristic regions; adding the reference microorganisms and exogenous nucleic acid into the to-be-measured sample, and extracting nucleic acid of the microorganisms in the to-be-measured sample; amplifying the nucleic acid of the microorganisms by use of the designed multi-amplification primers so as to obtain characteristic sequencing fragments; qualitatively and quantitatively analyzing the microorganisms in the to-be-measured sample by use of the characteristic sequencing fragments. The qualitative and quantitative detection method is free from preculture and proliferation of microorganisms, is capable of performing high-flux, high-accuracy and high-resolution detection on multiple types of known microorganisms in the to-be-measured sample in a step, and is simple and rapid in detection process and standard in process.
Description
technical field [0001] The invention relates to the field of biotechnology, in particular to a qualitative and quantitative detection method for rice microorganisms. Background technique [0002] Rice microorganisms are the basis for rice disease identification and prediction, rice food safety and other applications, and qualitative and quantitative detection is very necessary. [0003] Existing rice microbial qualitative and quantitative detection technologies include morphological counting, chip detection, 16SrRNA sequencing, metagenomic sequencing and real-time quantitative PCR (Polymerase Chain Reaction, polymerase chain reaction). Morphological counting detection requires pre-cultivation of microorganisms, which takes a long time, non-culturable microorganisms cannot be detected, and only one type of microorganism can be detected at a time, the throughput is low, the sampling amount is limited when counting, and the results are rough, and it is impossible to classify th...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/68C12Q1/06C12Q1/04C12Q1/70
CPCC12Q1/6869C12Q2531/113C12Q2537/143C12Q2535/122
Inventor 翟文学李甜甜彭海周俊飞陈利红李论胡长峰高利芬章伟雄张静卢龙陈斌李丽丽
Owner INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com