Probe composition group for detecting Pseudomonas bacteria
A technology of pseudomonad and composition, applied in the field of probe composition, can solve the problems of difficult repeatability and high price, and achieve the effect of low equipment requirements, low price and improved stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
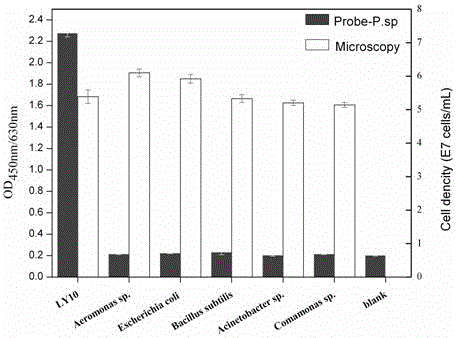
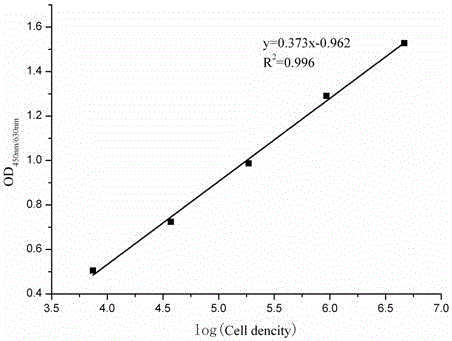
[0028] In order to understand the technical content of the present invention more clearly, the following examples are given in detail for the purpose of better understanding the content of the present invention rather than limiting the protection scope of the present invention. In this example, specific probes were used to detect nitrogen and phosphorus removal bacteria in urban sewage treatment, and their standard curves were prepared to verify their specificity.
[0029] 1. Design and synthesis of probes for specific regions of 16rRNA in Pseudomonas bacteria:
[0030] Comparing the 16rRNA sequence of Pseudomonas bacteria with other sewage bacteria sequences, it is found that it can be complementary to all species of Pseudomonas genus, but has no homology with other main sewage bacteria nucleotides, and it is determined as a characteristic region, and the design The analysis probes are:
[0031] SEQ ID NO: 1:
[0032] 5'-CGGCTAGTTGACATCGTTTACGGCGTGGACTACCAGGGTATCTAATCCTGTTT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com