Analysis method of unexpected effect of transgenosis plants
A technology of unexpected effects and transgenic plants, applied in the field of genetic engineering, can solve problems such as the methylation status of endogenous genes with low homology, and achieve important economic and social benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Extraction and Bisulfite Treatment of Genomic DNA
[0026] 1. Extraction of genomic DNA from transgenic plants
[0027] Under the same cultivation conditions, wild-type Arabidopsis (WT), Arabidopsis fhy3 gene deletion mutant (fhy3), and Arabidopsis thaliana transfected with fhy3 and GUS genes ( FHY3-GUS) seedlings, wild-type cucumber (WT2) and GUS-transgenic cucumber (GUS) seedlings, the genomic DNA of their leaves were extracted using the traditional CTAB method.
[0028] 2. Bisulfite treatment of genomic DNA
[0029] Arabidopsis and cucumber genomic DNA were subjected to bisulfite treatment according to the EZDNAMethylationGold kit (ZymoResearch) instructions.
Embodiment 2
[0030] Example 2: Cloning and sequencing of PCR amplification products
[0031] 1. PCR amplification of endogenous genes
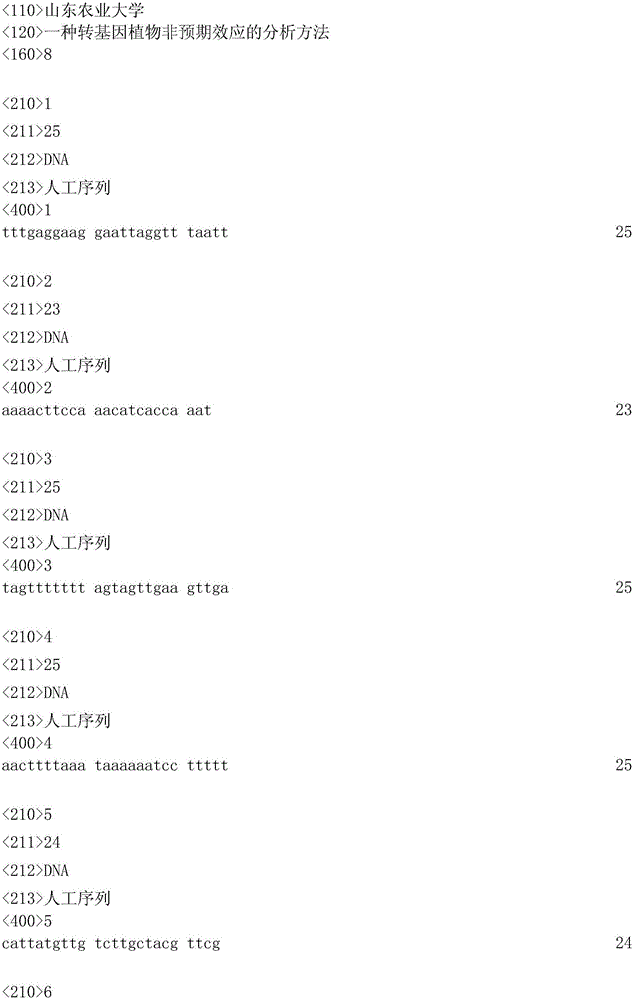
[0032] Select Arabidopsis thaliana endogenous gene AT4G16835 and cucumber endogenous gene Csa026131 that are not homologous to fhy3 and GUS genes, and use the online MethyPrimer software to design primers for a CpG island in its coding region as follows: The forward primer of the AT4G16835 gene is 5'- TTTGAGGAAGGAATTAGGTTTAATT-3', its nucleotide sequence is shown in SEQNo.1, the reverse primer is 5'-AAAACTTCCAAACATCACCAAAT-3', its nucleotide sequence is shown in SEQNo.2; the forward primer of Csa026131 gene is 5' -TAGTTTTTTTAGTAGTTGAAGTTGA-3', the nucleotide sequence of which is shown in SEQNo.3, and the reverse primer is 5'-AACTTTTAAATAAAAAATCCTTTTT-3', the nucleotide sequence of which is shown in SEQNo.4.
[0033] PCR amplification was performed using bisulfite-treated genomic DNA as a template. 20μL PCR reaction system includes: 0.4μL MgCl 2 (25mM), ...
Embodiment 3
[0044] Embodiment 3: Analysis of unexpected effects of transgenic plants
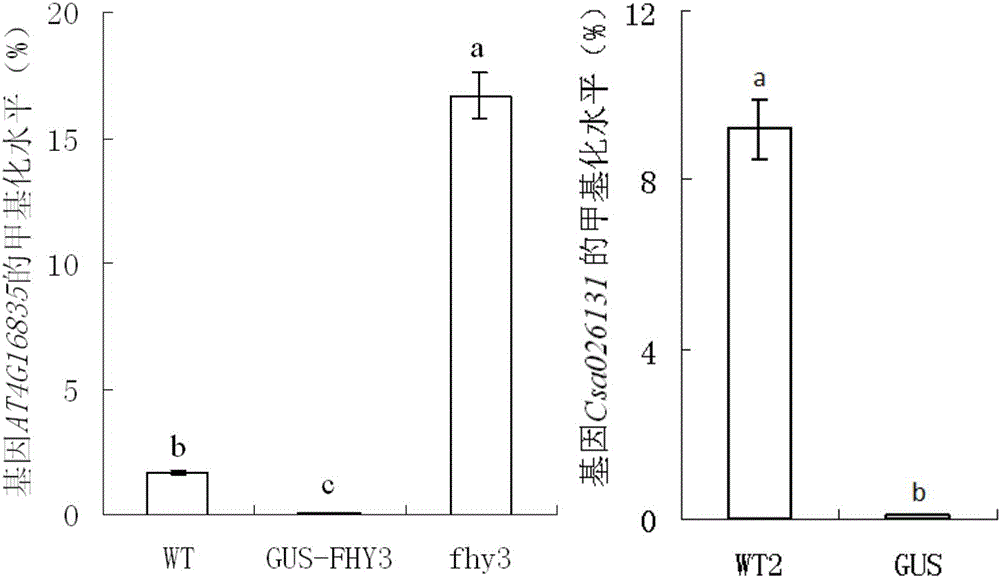
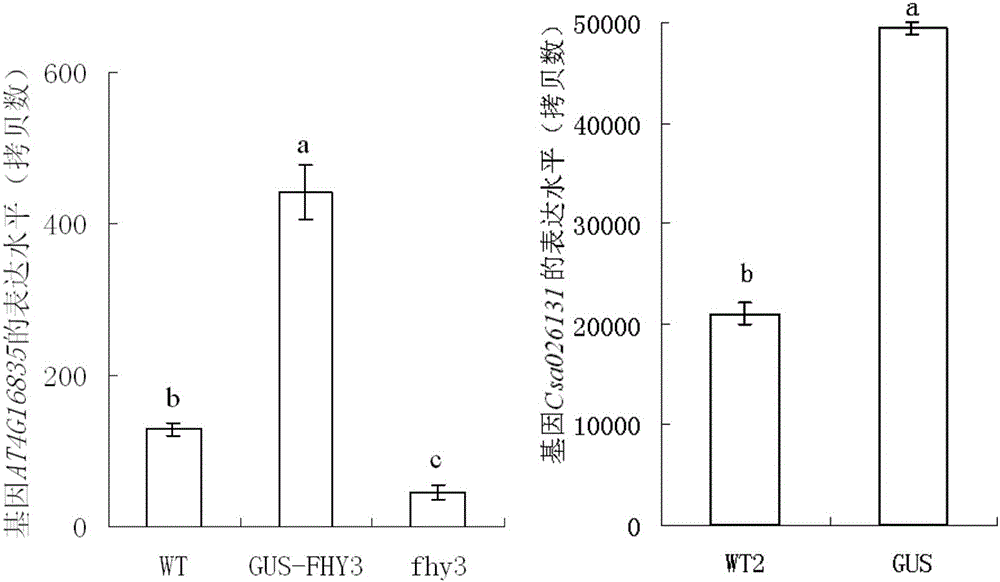
[0045] 1. According to the sequencing results of the PCR amplification products, calculate the ratio of unmethylated cytosine to the total cytosine in each clone, and obtain the average methylation level of each test material (20 clones). The experimental data are expressed in the form of mean ± standard error, using the statistical software SPSS13.0, through the one-way analysis of variance and TukeyHSD test for significant difference analysis, when P<0.05, there is a significant difference between the data.
[0046] Such as figure 1 showed that the average methylation level of endogenous gene AT4G16835 was higher in Arabidopsis fhy3 gene deletion mutant (fhy3), lower in wild-type Arabidopsis (WT), and was higher in fhy3 gene deletion mutants. The average methylation level was the lowest in the receptor, transfhy3 and GUS transgenic Arabidopsis (FHY3-GUS). Therefore, although the endogenous gene AT4G...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com