Method for fast identifying type of gene alternative splicing and application thereof
A gene and variable technology, applied in the field of identifying the alternative splicing type of a specific gene in eukaryotes at a specific time in a specific tissue, can solve the problem of inability to quickly determine the alternative splicing type of a gene, and achieve short identification time and experimental steps. Less, the result is easy to interpret
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
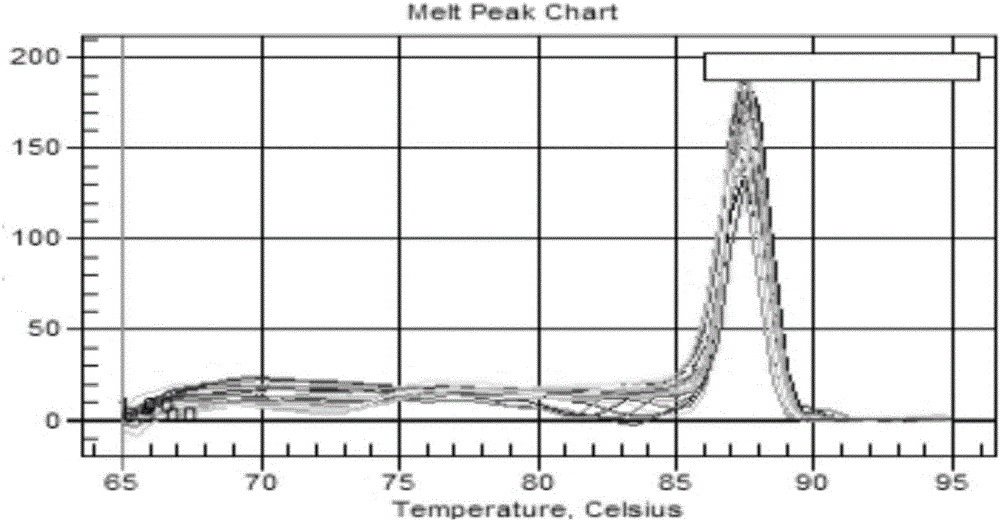
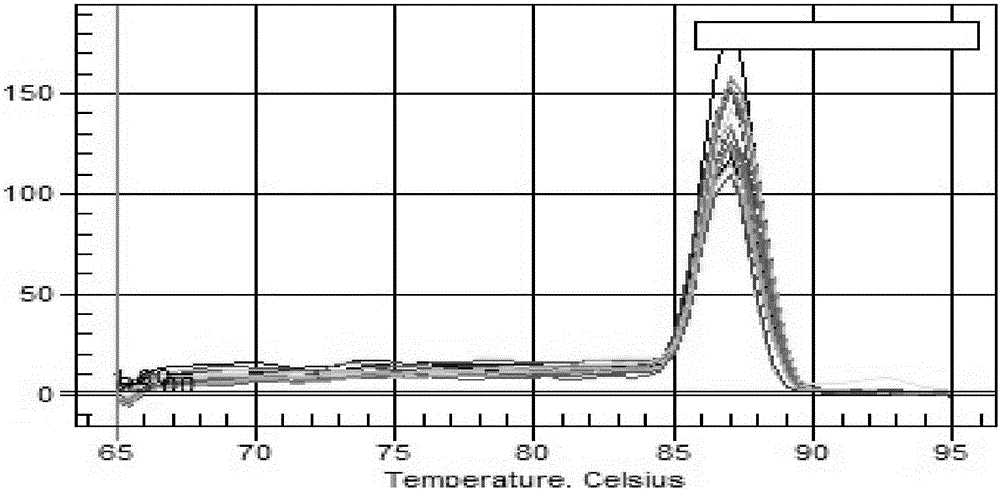
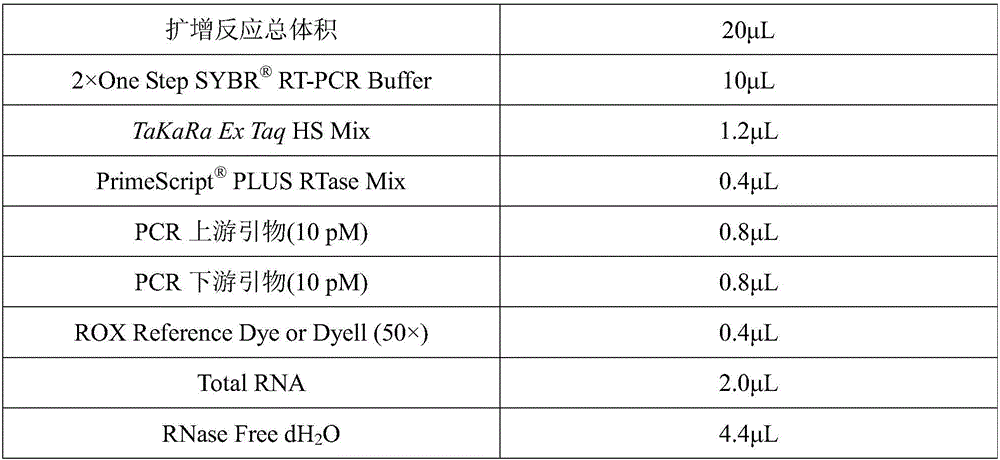
Image
Examples
Embodiment 1
[0056] Example 1 Rapid identification of the alternative splicing type of the test chicken MHC I gene
[0057] Take the detection of the alternative splicing type of the chicken MHC I gene as an example: all the alternative splicing types of the chicken MHC I gene obtained from the GenBank database and literature retrieval and / or other methods (such as through other known experimental methods), and then design a suitable The primer pair is used to amplify all the alternative splicing types of the chicken MHC I gene by real-time fluorescent quantitative PCR, and their melting temperatures are obtained respectively, so that the standard melting temperature curve library of the gene is constructed, and the chicken The steps of MHCⅠgene standard melting temperature curve library are as follows:
[0058] (1) Search and obtain all alternative splicing types of chicken MHC I gene from GenBank database and literature; use Oligo6.31 or PrimerPremier5.0 software to design primer pairs, ...
Embodiment 2
[0078] Example 2 Rapid identification of the alternative splicing type of the test pig TLR4 gene
[0079] The difference between this example and Example 1 is that the gene to be tested is domestic pig TLR4 gene, and the primer pair is: upstream primer 5'-GACCCTTGCGTGCAG-3'; downstream primer 5'-CTCTGGATAGGATTTCC-3'. There are three gene alternative splicing types in this gene: TLR4 complete type, TLR4 exon 2 complete deletion type, and TLR4 exon 3 partial deletion type; the sequences of each gene are as follows:
[0080] (1) Complete gene sequence of TLR4
[0081] 5'-GACCCTTGCGTGCAGGTGGTTCCTAACATTAGTTACCAATGCATGGAGCTGAATTTCTACAAAATCCCTGACAACATCCCCACATCAGTCAAGATACTGGACCTGAGCTTTAACTACCTGAGTCATTTAGACAGCAATAGCTTCTCCAGCTTTCCAGAACTGCAGGTGCTGGATTTATCCAGATGTGAAATTCAGACAATTGACGATGATGCATATCAGGGCCTAAATTACCTTTCCACCTTGACACTGACGGGAAATCCTATCCAGAG-3';
[0082] (2) Partial deletion gene sequence of exon 3 of TLR4
[0083] 5'-GACCCTTTGCGTGCAGGTGGTTCCTAACATTAGTTACCAATGCATGGAGCTGAATTTCTACAAAAT...
Embodiment 3
[0087] Example 3 Rapid Identification Test Alternative Splicing Type of Longan MEE70 / MSll Gene
[0088] The difference between this example and Example 1 is that the gene to be tested is the longan MEE70 / MS11 gene, and the primer pair is: upstream primer 5'-CGGTTGTTGCTCATC-3'; downstream primer 5'-CTGGGCTTCCATATC-3'. There are two types of gene alternative splicing in this gene: MEE70 / MS11 complete type, MEE70 / MS11 exon 4-7 deletion type; wherein, each gene sequence is as follows: (1) MEE70 / MS11 complete type gene sequence
[0089] 5'-CGGTTGTTGCTCATCAGAGTGAGGTTAACTGCTTAGCATTCAATCCCTTTAATGAATGGATTTTGGCGACTGGGTCTACTGATAAGATGGTTAAGTTATTTGATTTGCGCAAGATTAGCACGGCACTTCACACATTTAGTCACAAGGAGGAAGTTTTCCAAGTTGGATGGGACCCAAAGAGTGAGACTATTTTAGCATCTTGTTGTCTTGGTAGAAGGTTGAAGGTGTGGGATCTTAGGAGGATCGATGAGGAACAGACACTAGAGGATGCCGAAGATGGTCCACCAGAGTTGCTTTTTACTCATGGTGGTCACACGAGTAAAATCTCAGATTTTTCATGGAACCCATGTGAAGATTGGGTTATTGCTAGCGTAGCGGAAGATAATATTCTTCAAATATGGCAGATGGCAGAGAACATTTACCATGATGAAGATGATTTACCTGGAGATGA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com