Use of PARP inhibitors to treat breast or ovarian cancer patients showing a loss of heterozygosity
An inhibitor, breast cancer technology, applied in the field of genome-wide LO in ovarian cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
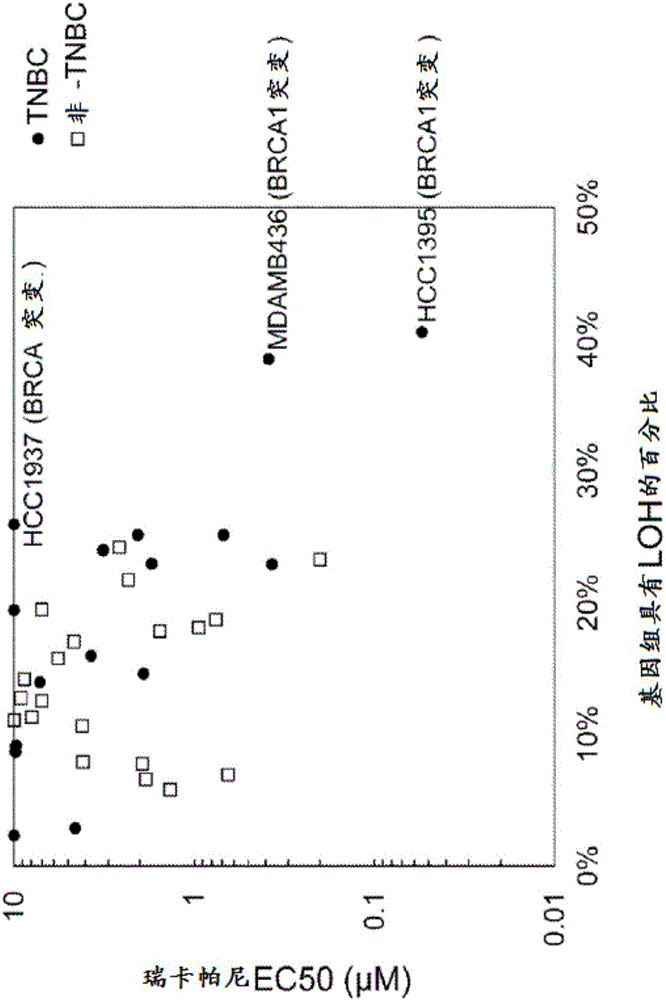
[0052] Ricaparib-sensitive breast cancer cells exhibit genomic LOH
[0053] Ricaparib-sensitive cells
[0054]Sensitivity data for ricaparib in a large panel of human cancer cell lines were generated using a high-throughput growth inhibition assay. Briefly, place cells between 5 and 20x 10 3 Cells were seeded in 24-well tissue culture plates. Ricaparib was treated at concentrations ranging from 0.005 to 10 μΜ. On days 1 and 6 of ricaparib treatment, viable cells were counted using a Beckman Coulter Z2 particle counter. Growth inhibition was calculated as a function of the number of inhibited generations in the presence of ricaparib relative to the number of generations over the same time course in the absence of ricaparib. Dose response curves were generated and half maximal effective concentration (EC50) values for growth inhibition were calculated for each cell line. Some of the most sensitive cell lines seen in high throughput screens were breast cancer cell lines (T...
Embodiment 2
[0069] Example 2 -Ovarian cancer patients whose tumors exhibit genomic LOH benefit from platinum-based therapy
[0070] high-grade serous ovarian neoplasms
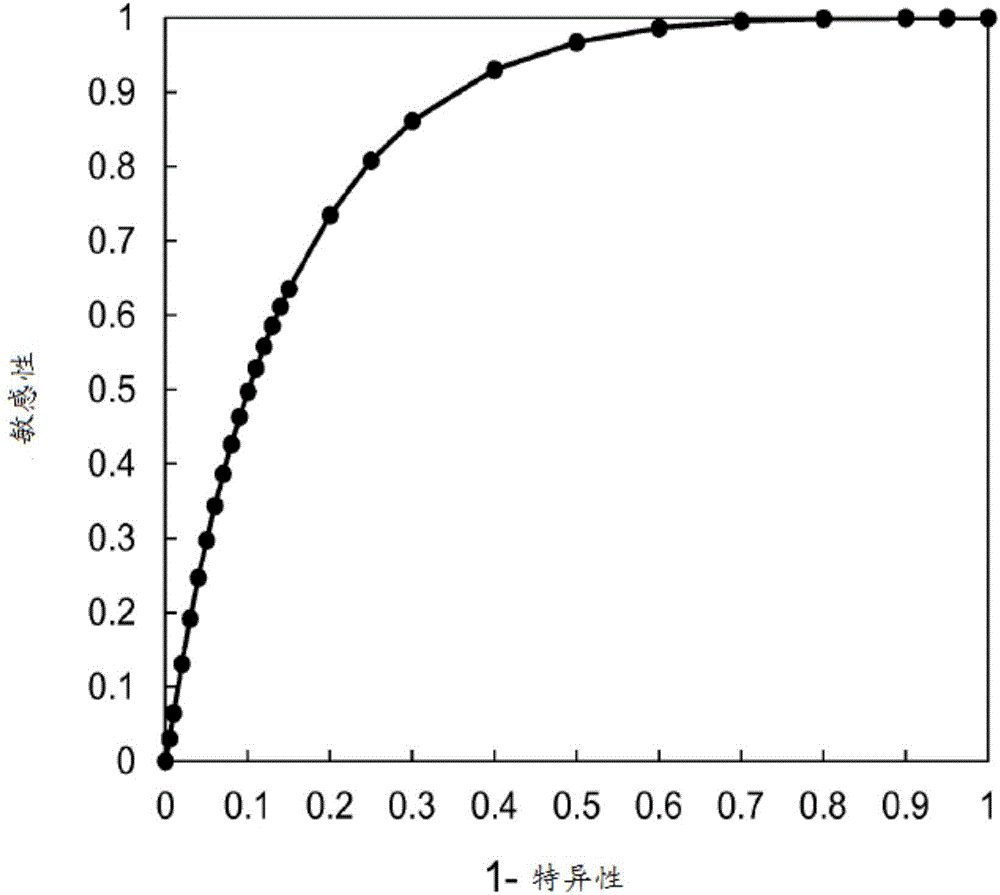
[0071] The Cancer Genome Atlas (TCGA) project performed genomic analyzes of 316 high-grade serous ovarian tumors (Cancer Genome Atlas Research Network. Integrated genomic analyzes of ovarian carcinomas. Nature 2011;474:609-15). Samples were collected from patients newly diagnosed with ovarian serous adenocarcinoma who were undergoing surgical resection and had not received prior treatment. Patients were then treated with platinum-based chemotherapy as with standard therapy, and overall survival (interval from date of initial surgical resection to date of last known association or death) was recorded. Patients with a platinum-free interval (interval from date of last primary platinum therapy to date of progression) of 6 months or greater were identified as platinum-sensitive. Next-generation sequencing of tumors to ide...
Embodiment 3
[0083] Example 3 – Ovarian cancer patients whose tumors exhibit genomic LOH benefit from treatment with ricaparib
[0084] Next-generation sequencing of ovarian tumors from patients
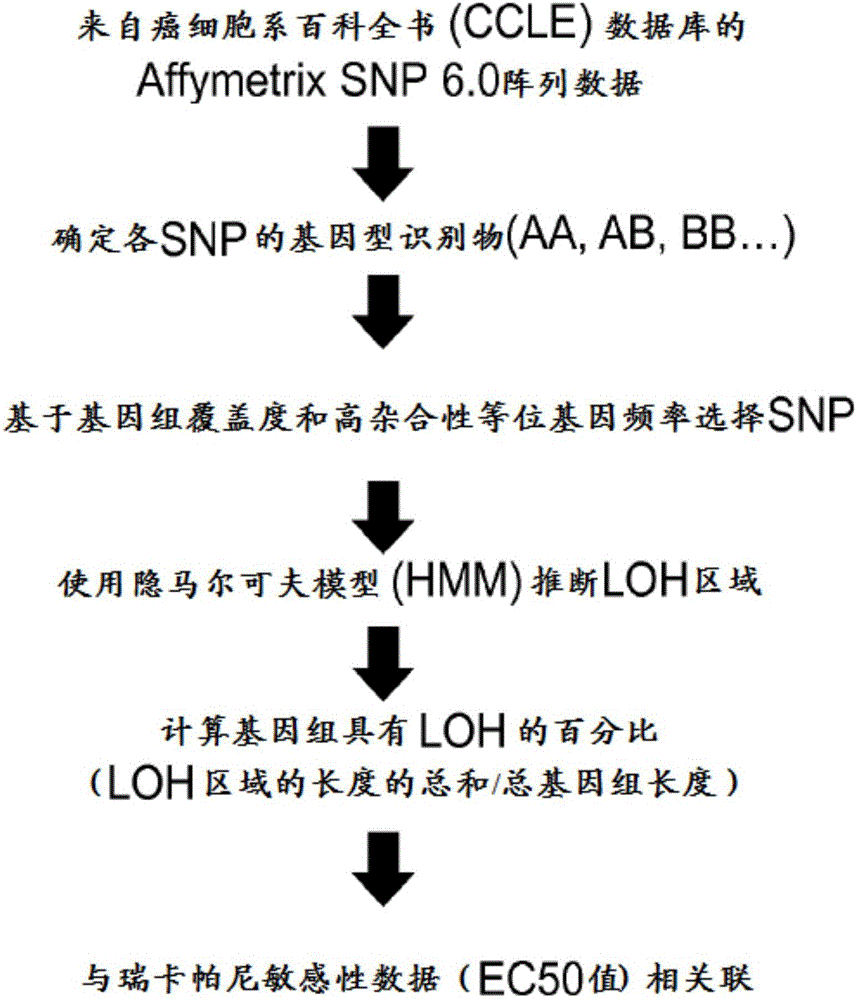
[0085] Archival tumor tissue samples from patients are optionally collected for genomic analysis. To determine the maximum tolerated dose and recommended Phase II dose, patients were placed in dose escalation cohorts for continuous daily oral administration of ricaparib. The antitumor activity of ricaparib was evaluated based on the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. In addition, the concentration of cancer antigen-125 (CA-125) in blood was measured as a biomarker of ovarian cancer. Local BRCA1 / 2 test results are based on sequencing of the BRCA1 / 2 gene from a blood sample (peripheral blood mononuclear cells).
[0086] LOH analysis
[0087] A high-level overview of the bioinformatics analysis workflow is given in Figure 10 middle. Briefly, formalin-fixed pa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com