Preparation method of molecular tag
A molecular tag and sequence technology, applied in the field of DNA variation detection, can solve the problems of data unavailability, long operation cycle, and lower data efficiency, and achieve the effects of improving reaction rate, evaluating preference, and reducing waste
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
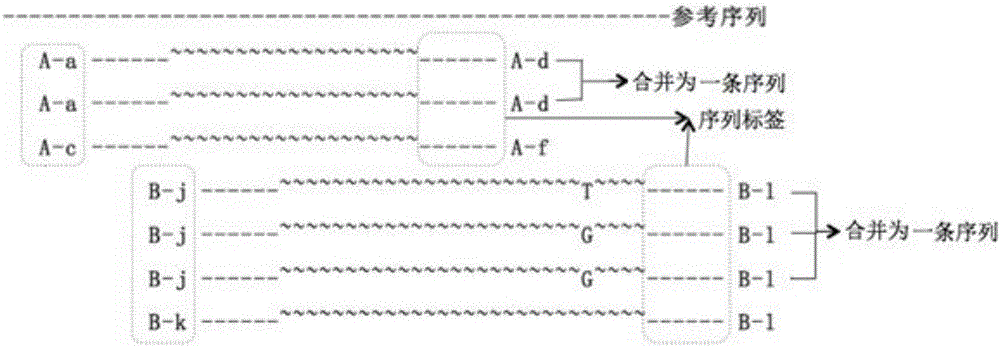
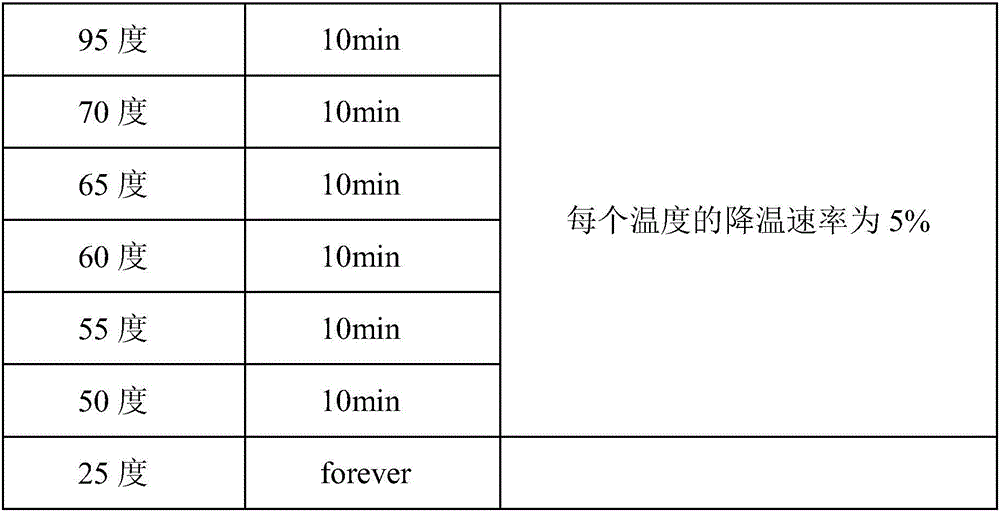
Examples
Embodiment 1
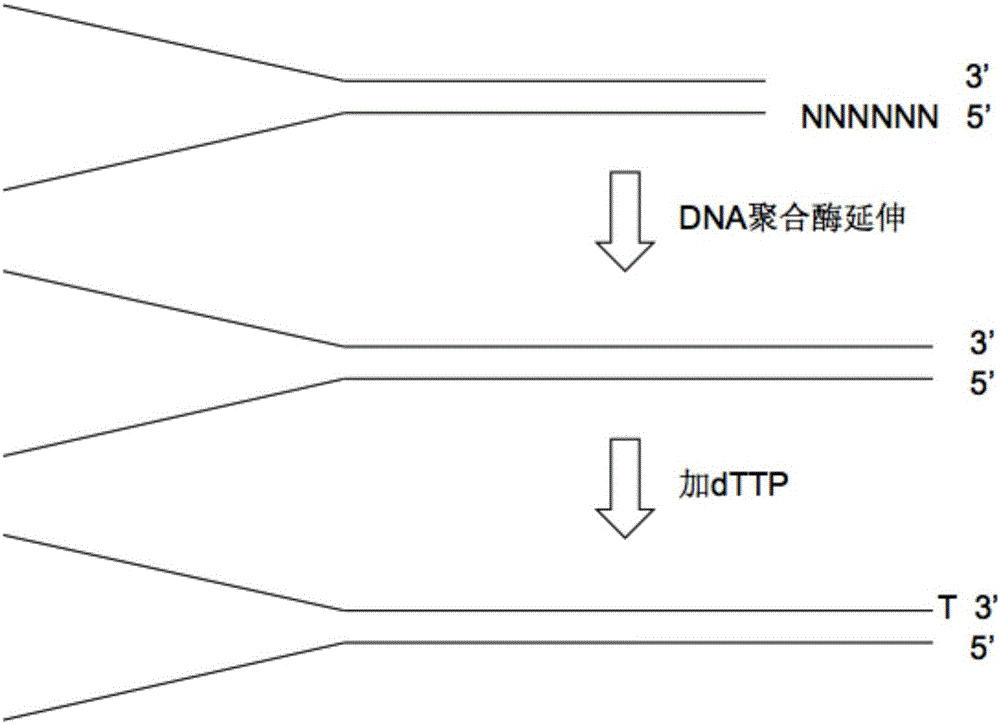
[0037] 1. Synthesis and annealing of the adapter
[0038](1) Take a tube of sequencing adapter DNA whose sequences are 5'-TACACTCTTTCCCTACACGACGCTCTTCCGATCT-3' and 5'-TCTTTCTACAGTCANNNNNNAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3', where NNNNNN in the sequence is a 6bp random barcode, where N means A / T / G / Any base in C will do. That is, the sequences contained in the adapter DNA are at most 4 to the 6th power, and the difference of these sequences depends on what base N is. Centrifuge at 20,000 g for 10 min at 4°C to ensure that the adapter powder gathers at the bottom of the tube, and the labels are adapter-1 and adapter-2 respectively.
[0039] (2) Open the cap of the tube. At this time, be careful not to let the adapter powder float out of the tube. Add Nuclease-Free water (nuclease-free water) to adapter-1 and adapter-2 to dissolve the powder to make the concentration 100nmol / mL, that is, 100μM; then store at -20°C, if not used for a long time, store at -80°C Save it.
[004...
Embodiment 2
[0073] 1. Synthesis and annealing of the adapter
[0074] (1) Take a tube of sequencing adapter DNA whose sequences are 5'-TACACTCTTTCCCTACACGACGCTCTTCCGATCT-3' and 5'-TCTTTCTACAGTCANNAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3', where NN in the sequence is a 2bp random barcode, where N represents A / T / G / Any base in C will do. Centrifuge at 20,000 g for 10 min at 4°C to ensure that the adapter powder gathers at the bottom of the tube, and the labels are adapter-1 and adapter-2 respectively.
[0075] (2) Open the cap of the tube. At this time, be careful not to let the adapter powder float out of the tube. Add Nuclease-Free water (nuclease-free water) to adapter-1 and adapter-2 to dissolve the powder to make the concentration 100nmol / mL, that is, 100μM; then store at -20°C, if not used for a long time, store at -80°C Save it.
[0076] (3) Take 25 μL of adapter-1 (100 μM) and 25 μL of adapter-2 (100 μM) and mix them uniformly to obtain 50 μL of a mother solution with a concentration...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com