Tomato spotted wilt resistance gene Sw-5b close linkage SNP site acquisition and marker development
A spot wilt, sw-5b technology, applied in the biological field, can solve the problems of low detection efficiency, affecting the breeding process, difficult breeding of disease-resistant varieties, etc., and achieves the effects of high detection efficiency and simple detection method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
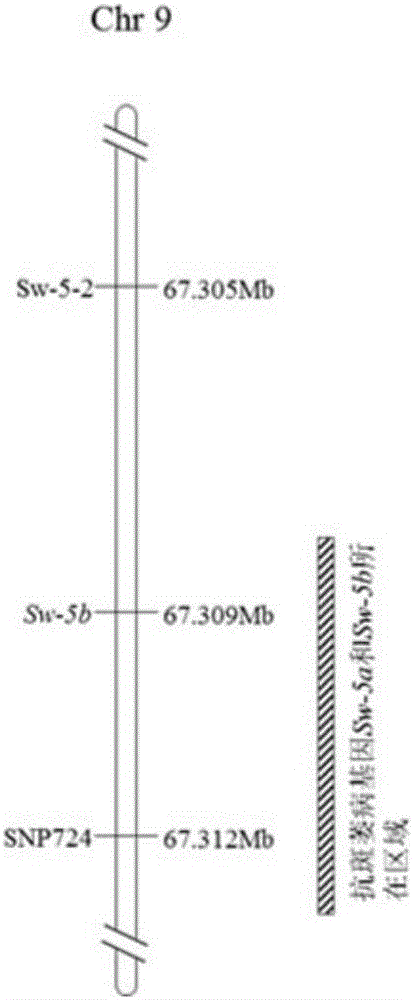
[0037] Example 1. Acquisition and identification of closely linked SNP sites and SNP markers with tomato spotted wilt resistance Sw-5b gene
[0038] After a large number of sequence analysis and discovery of SNP sites by the inventors, and correlation analysis between SNP sites and multiple traits, a SNP site related to tomato spotted wilt resistance was found, named SNP724.
[0039] The SNP724 marker sequence is as follows, and the polymorphic site is the 59th nucleotide of the following sequence 1, which is A or G;
[0040] 5'-CTACTTTTTTCCAATGGATAAAGCTTTGGAATGGAATCATGGACACAACTGGAGTTATT[A / G]CTATATATCTTTGAAACTTGTACAAGTAAAATTTGAAGAATGACTGTTG-3' (SEQ ID NO: 1)
[0041] According to SNP724 primer design, pre-experimental comparison effect, select the specific primer combination of SNP724 site:
[0042] FP1SNP724: 5'-GAAGGTCGGAGTCAACGGATTCATGGACACAACTGGAGTTATTA-3' (SEQ ID NO: 2)
[0043] FP2SNP724: 5'-GAAGGTGACCAAGTTCATGCTCATGGACACAACTGGAGTTATTG-3' (SEQ ID NO: 3)
[0044] RPSNP...
Embodiment 2
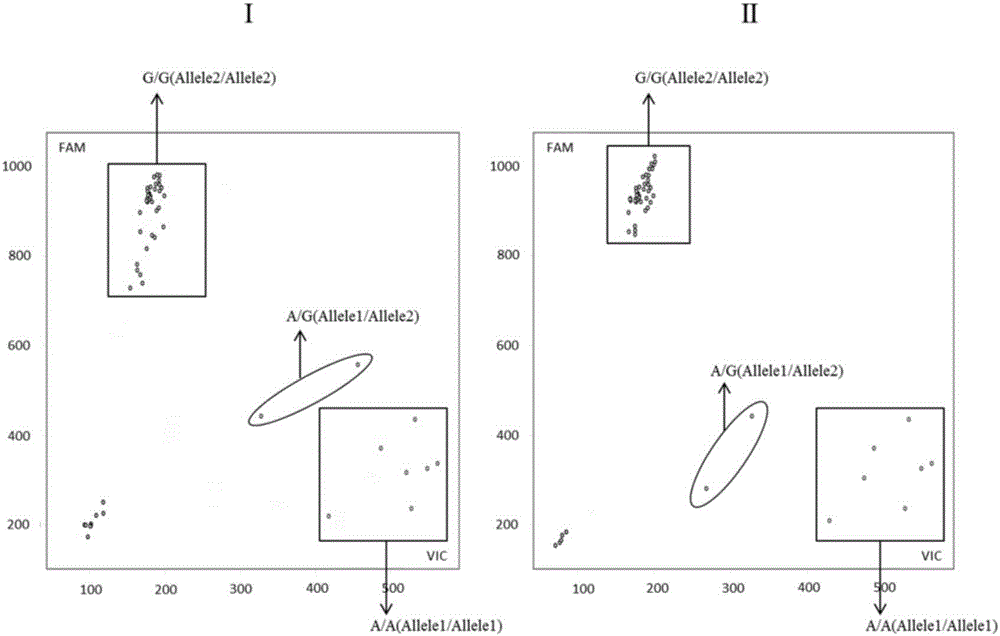
[0046] Example 2, the application of SNP sites and their specific primer combinations in identifying whether tomato contains the Sw-5b gene for resistance to spotted wilt
[0047] 1. Identify whether tomato contains the spotted wilt resistance Sw-5b gene
[0048] 1. Genomic DNA extraction
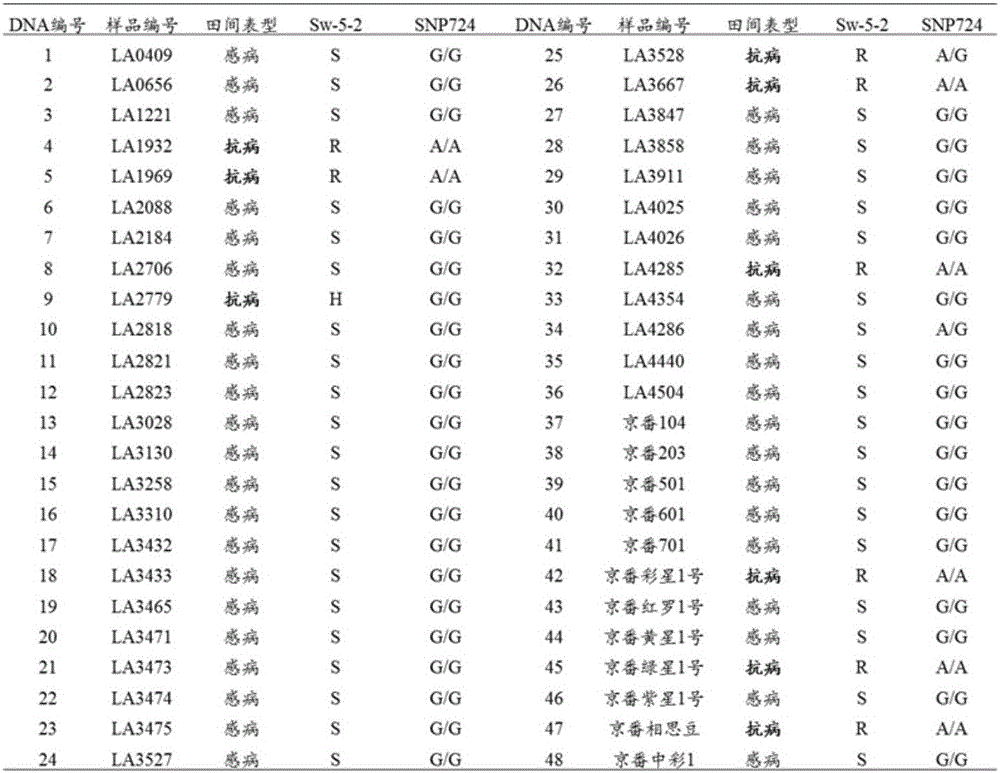
[0049] Genomic DNA of 48 tomato materials shown in Table 1 was extracted.
[0050] Table 1 Identification table of 48 tomato material phenotypes and marker genotypes
[0051]
[0052] Note: 1. The samples with DNA number 1-36 in the table are registered samples of Tomato Germplasm Resource Center (Tomato GeneticsResource Center http: / / tgrc.ucdavis.edu / index.aspx); the samples with DNA number 37-48 are commodity type;
[0053] 2. In column Sw-5-2, 'R' indicates the homozygous resistant genotype of spotted wilt, 'S' indicates the homozygous susceptible genotype of spotted wilt, and 'H' indicates the heterozygous resistant gene of spotted wilt type;
[0054] 3. In the SNP724 column, 'A...
Embodiment 3
[0071] Example 3, Application of SNP724 site and its specific primer combination in identification of resistance to spotted wilt of tomato
[0072] 1. Phenotypic detection of tomato resistance to spotted wilt
[0073] The 48 tomato materials from different sources shown in Table 1 were selected, and after planting in the field, the phenotype observation was carried out. 10 plants were planted in each material, and the disease resistance phenotype statistics were carried out during the high-incidence period of spotted wilt. If there were more than 7 disease-resistant plants in each material, this material was recorded as disease-resistant. The statistical results are shown in Table 1.
[0074] 2. Identification of tomato resistance to spotted wilt by SNP loci and their specific primer combinations
[0075] 1. Obtaining genomic DNA
[0076] Genomic DNA of 48 tomatoes from different sources was extracted.
[0077] 2. PCR amplification
[0078] The above genomic DNA was amplif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com