Extracting method for metagenome DNA (Deoxyribonucleic Acid)
An extraction method and metagenomics technology, applied in the direction of recombinant DNA technology, DNA preparation, etc., can solve the problem that host cells cannot be removed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Metagenomic DNA extraction of stool samples
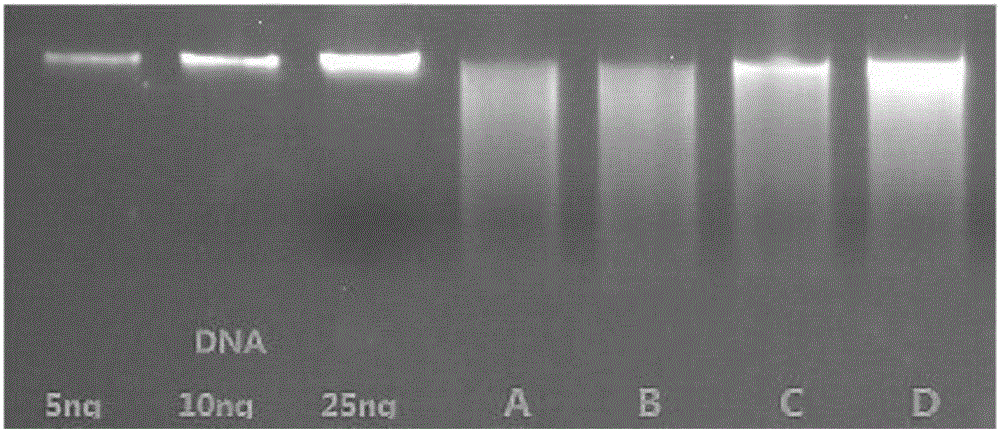
[0026] Take two clean 5ml centrifuge tubes, weigh 1g of two different fecal samples No. 1 and No. 2 into the centrifuge tubes, add 1.6ml of Buffer SLX Mlus to the two tubes, shake and mix evenly. Weigh 500 mg of glassbeads into four 2 mL centrifuge tubes numbered A, B, C, and D, respectively, and then add 5 μL of proteinase K at a concentration of 10 mg / mL to the centrifuge tubes numbered A and B, respectively. Then add 1 mL of No. 1 stool sample to centrifuge tubes numbered A and C, respectively, and add 1 mL of No. 2 stool sample to centrifuge tubes numbered B and D respectively. The components in the centrifuge tubes numbered A, B, C, and D (hereinafter referred to as tubes A, B, C and D) are shown in Table 1:
[0027]
A
B
C
D
1
2
1
2
buffer
1.6mL
1.6mL
1.6mL
1.6mL
5μL
5μL
0
0
glass beads
500...
Embodiment 2
[0039] Example 2 Metagenomic DNA extraction of blood samples
[0040] Two different blood samples were taken to extract metagenomic DNA according to the method of Example 1. The blood samples in tubes A and B were added with 5 μL proteinase K at a concentration of 15 mg / mL, heated at 60 °C for 20 min to lyse the host cells by proteinase K, and then heated at 80 °C for 20 min to inactivate proteinase K, and then followed Extract metagenomic DNA according to the instructions of the kit. The blood samples in tubes C and D were directly extracted for metagenomic DNA according to the operating steps of the kit instructions.
[0041] The above metagenomic DNA from blood samples was detected by 2% agarose gel electrophoresis. figure 1 similar. Then, library construction and sequencing analysis were performed. The results are shown in Table 4. The content of host DNA in metagenomic DNA without proteinase K heat treatment was about 6 times that after proteinase K heat treatment.
...
Embodiment 3
[0044] Example 3 Metagenomic DNA extraction of tissue samples
[0045] Two different human intestinal tissue samples were taken, and metagenomic DNA was extracted according to the method of Example 1. The intestinal tissue samples in tubes A and B were added with 5 μL of proteinase K at a concentration of 20 mg / mL, heated at 55 °C for 30 minutes to lyse the host cells by proteinase K, and then heated at 90 °C for 15 minutes to inactivate proteinase K, then Then, extract metagenomic DNA according to the operating steps of the kit instructions. The intestinal tissue samples in tubes C and D were directly extracted from metagenomic DNA according to the instructions of the kit.
[0046] The above metagenomic DNA from intestinal tissue samples was detected by 2% agarose gel electrophoresis. figure 1 similar. Then, library construction and sequencing analysis were performed. The results are shown in Table 5. The content of host DNA in metagenomic DNA without proteinase K heat tre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com