Host cell modification with artificial endosymbionts
An artificial endosymbiont and host cell technology, applied in the direction of genetically modified cells, fused cells, cells modified by introducing foreign genetic material, etc., can solve the problems of high complexity and complexity of eukaryotic cells, slow growth of bacterial cells, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0151] Example 1: Translocation of proteins from artificial endosymbionts into host cells using the type I secretion system
[0152] The type I secretion system (T1SS) in Gram-negative bacteria can be used to export a variety of proteins (their cognate substrates) with different sizes and diverse functions. The MTB genome encodes the T1SS gene (Matsunaga, T. et al., "Completegenome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. AMB-1", DNA Res. 12:157-66, 2005; incorporated herein by reference in its entirety ).
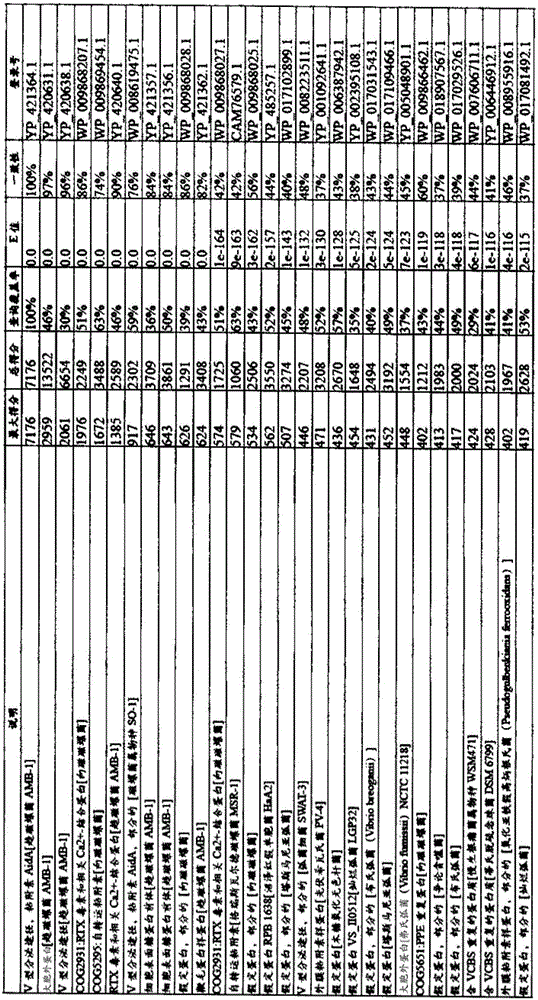
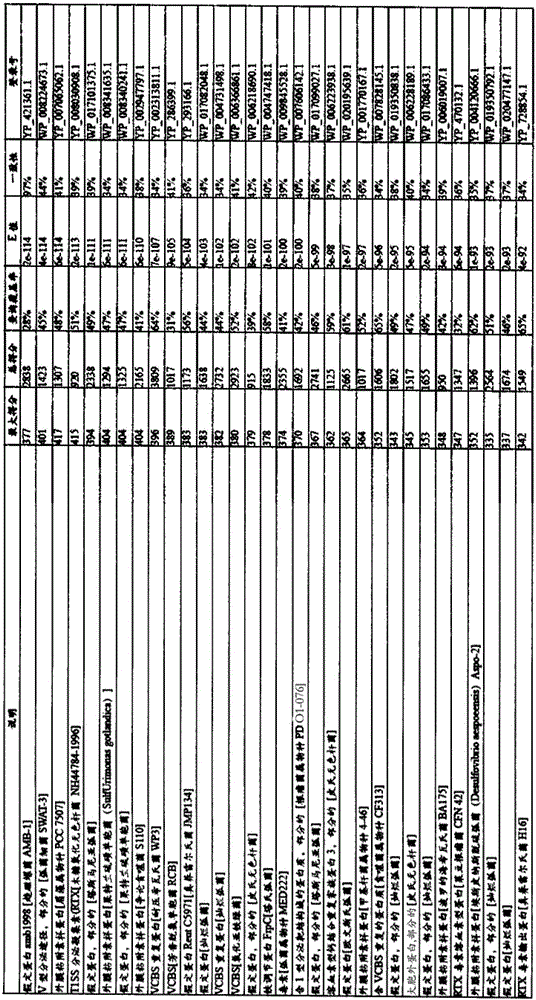
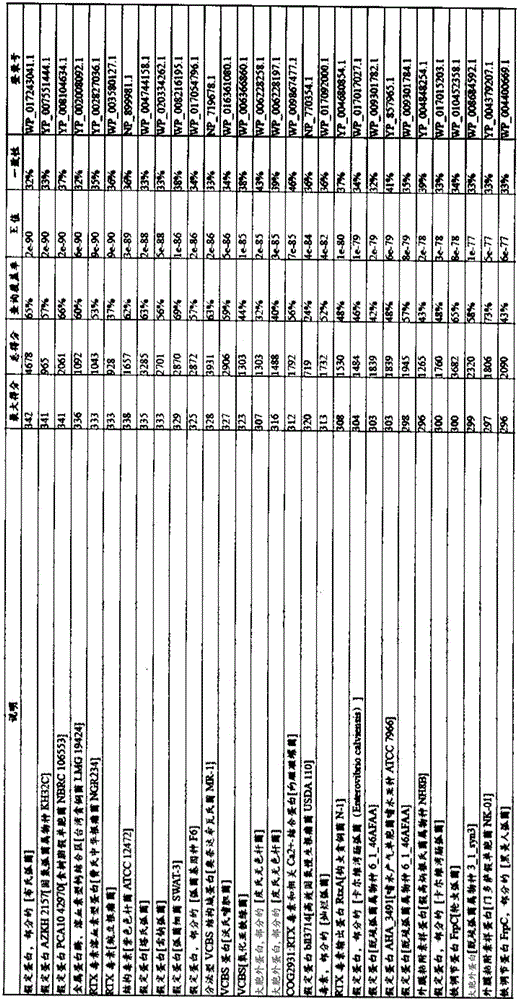
[0153] The target protein, green fluorescent protein (GFP), or red fluorescent protein (RFP), was N-terminally fused to YP_420640 (RTX toxin and related Ca 2+ binding protein), YP_423419 (RTX toxin and related Ca 2+ binding protein), YP_422785 (amb3422), or the C-terminal 200 amino acids of other MTB T1SS substrates. Alternatively, the proteins in MTB-YP_420502, YP_420631.1, YP_420638.1, YP_420640.1, YP_421364.1, YP_422662.1, YP...
example 2
[0159] Example 2: Translocation of proteins from artificial endosymbionts into host cells using the type IV secretion system
[0160] The CagA protein was engineered by replacing the last 20 amino acids with 24 amino acids from the C-terminal end of the RSF1010MobA protein (residues 684-709), which is secreted by the T4SS system. Translocation of CagA-MobA into host cells was monitored by observing the hummingbird phenotype caused by CagA in the host cells. The hummingbird phenotype is disclosed in Segal et al. by the spreading and elongated growth of cells, the presence of lamellipodia (thin sheets of actin present at cell edges), and filopodia-like structures (containing actin Finger-like projections characterize tight bundles of protein filaments (see Segal, E.D. et al., "Altered States: Involvement of Phosphorylated Caga in the Induction of Host Cellular Growth Changes by Helicobacter Pylori", Proc Natl Acad Sci. USA 96(25):14559 -64, 1999).
[0161] The hummingbird phen...
example 3
[0168] Example 3: Translocation of Nucleic Acids from Artificial Endosymbionts into Host Cells Using the Type IV Secretion System
[0169] The CagA4-type secretion system (T4SS) can be used to transfer plasmid DNA from artificial endosymbionts into mammalian host cells. Sequence analysis revealed that components of the T4SS system are present in MTB.
[0170] Plasmid pBBR-MSC was engineered to contain (oriT+trwABC) for plasmid transfer via the T4SS system, and the plasmid was engineered to contain expression of the target protein encoded under the control of the HCMVIE1 promoter-enhancer-first intron box. The target protein is a selectable marker such as DHFR or glutamate synthetase, or a reporter protein such as GFP, or a transcription factor such as cMyc.
[0171] Mammalian selectable markers can be used as target genes. These genes include the puromycin N-acetyltransferase gene for puromycin resistance; blasticidin S deaminase for blasticidin S resistance; and the amino ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com