Milbemycin positive regulation gene milR and overexpressed genetic engineering bacterium, preparation method and application thereof
A technology of milbemycin and genetically engineered bacteria, applied in genetic engineering, botany equipment and methods, methods based on microorganisms, etc., can solve problems such as failure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The research of embodiment 1milR gene function
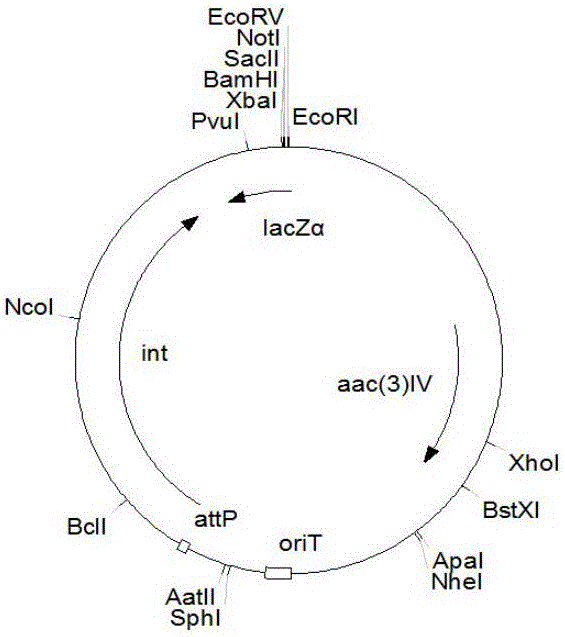
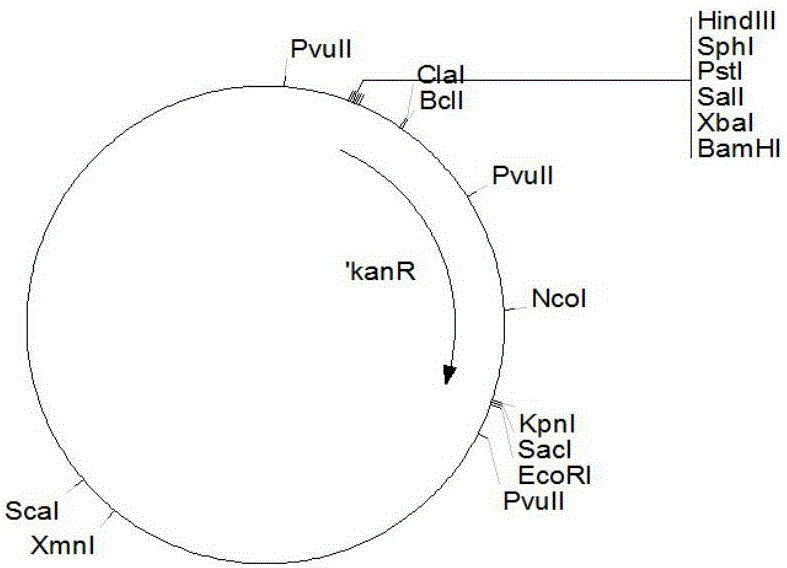
[0031] 1. Construction of recombinant plasmids for gene disruption
[0032] Using primer RD-LF (5'CG GAATTCCAGCGACACCATCACCGAGAACA3', the underline is the EcoRI site) and RD-LR (5'CGG GGTACC GACTGAGCGAGTCGGAAATT3', the underline is the KpnI site) PCR amplified from the Streptomyces bingchengsis X-7 genome to obtain the homologous recombination left arm fragment of the milR gene. The total PCR reaction system is 100 μl, and the reaction conditions are: 94°C, 4min; 94°C, 1min, 68°C, 3min, 31 cycles; 68°C, 10min. After the PCR product was loaded and electrophoresed, the target band with a length of 2.2kb was recovered, and then the recovered fragment was digested with EcoRI and KpnI.
[0033] Using primer RD-RF (5'CG GGATCC CAGCAGTCGGCTCAGCAACG3', BamHI site is underlined) and RD-RR (5'GC TCTAGA CTCGAAATCCTTCTGGGTCAGGT3'), the underline is the XbaI site) PCR amplified from the Streptomyces bingchengsis X-7 genome to...
Embodiment 2
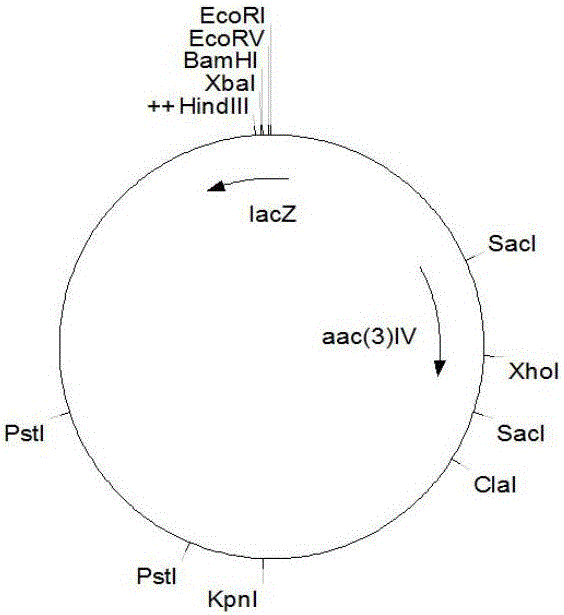
[0045] Embodiment 2, the construction of milbemycin high-yielding engineering strain
[0046] Transform E.coli ET12567 / pUZ8002 competent cells with the recombinant vector pSET152::milR used for complementation in Example 1 to obtain a single clone of E.coli ET12567 / pUZ8002 containing pSET152::milR, which was transferred into Streptomyces bingchenggensis BC- In the 120-4 strain, the process is to pick a single clone and inoculate it in the liquid LB medium with corresponding resistance (add 100 μg / ml apramycin and 25 μg / ml chloramphenicol during cultivation); cultivate overnight at 37°C; The overnight culture was inoculated in liquid LB medium supplemented with corresponding antibiotics at a ratio of 1:100, and cultivated at 37°C until OD600=0.4-0.6; centrifuged, collected bacteria, and washed the bacteria twice with an equal volume of liquid LB medium. Once, use 1 / 10 volume of 2YT to suspend; while washing E. coli cells, use 2×YT medium to collect Streptomyces spores, transfer...
Embodiment 3
[0049] The difference from Example 1 is that Streptomyces bingchengsis X-7 is replaced by Streptomyces bingchenggensis BC-120-4. Other steps are with embodiment 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com