Multiple PCR primer pair combination for bacterium determination and determination method
A technology of primer pairs and bacteria, which is applied in biochemical equipment and methods, recombinant DNA technology, measurement/inspection of microorganisms, etc., can solve the problems of inability to distinguish bacteria, and achieve the effect of efficient identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
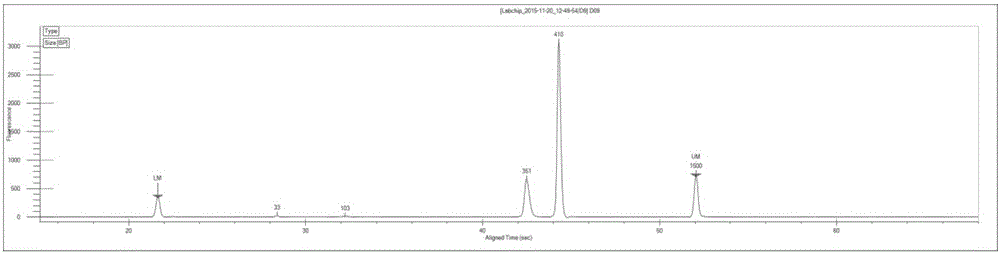
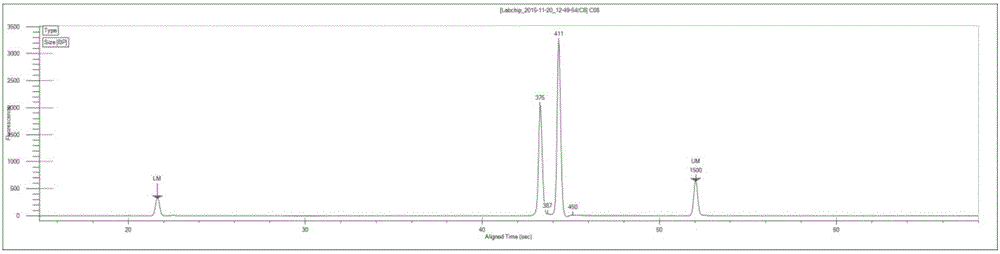
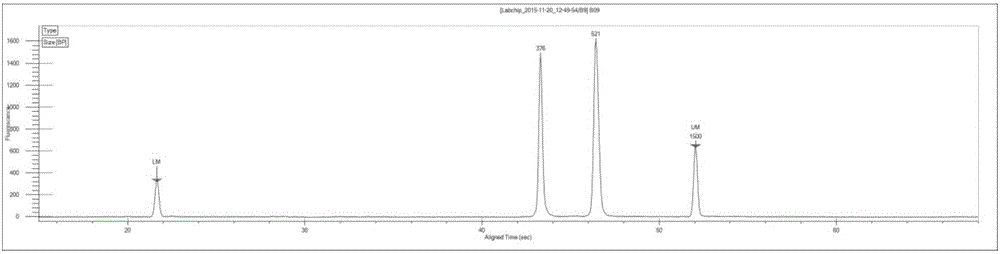
Image
Examples
Embodiment 1
[0052] Embodiment one: the design of primer pair combination
[0053] 1. Design of primer pairs
[0054] According to the conserved region sequence of 16S rDNA, primer pairs were designed to amplify 8 variable regions, among which, the V1 and V2 regions were amplified using the P1 primer pair, the V3 and V4 regions were amplified using the P3 primer pair, and the V5 and V6 regions were amplified using the P5 primers For amplification, the V7 and V8 regions were amplified using the P6 primer pair (Table 1a, 1b).
[0055] In Table 1a and 1b, the underlined primer sequence is the sequence that binds to the 16S rDNA conserved region, and the primer sequence that is not underlined is the sequence that is complementary to the sequencing general primer of the Illumina Miseq V1-V2 kit. Between the sequence complementary to the sequencing primer and the sequence combined with the 16S rDNA conserved region, different tag sequences can be added according to the final library sample numb...
Embodiment 2
[0066] Example 2: Screening of primer pair combinations
[0067] 1. Extract bacterial nucleic acid
[0068] 1) Use the DNA Purification from Blood or Body Fluids (Spin Protocol) (Qiagen Cat.NO.51306) kit to extract the genomic DNA of Legionella pneumophila (purchased from Beijing Beinan Chuanglian Biotechnology Research Institute).
[0069] 2) Quantify the DNA in the collection tube with a spectrophotometer. The requirements for DNA used for flux detection are: concentration: ≥10ng / μl; purity: OD260 / 280 of 1.8-2.0; total amount of nucleic acid: ≥1μg. Short-term storage at -20°C; long-term storage at -80°C.
[0070] 2. PCR amplification
[0071] 1) Synthesize each primer in the P1, P3, P5, and P6 primer pairs and dilute them into 5 μM use solution;
[0072] In this example, there is only one sample, so select one tag sequence, which is the index sequence "TGAACCTT" provided by the Illumina sequencing platform; that is, the sequences of each primer in the P1, P3, P5, and P6 ...
Embodiment 3
[0100] Embodiment three: Utilize primer pair combination to identify bacteria
[0101] 1. Strains
[0102] Select 6 strains (Clostridium difficile, Enterococcus faecium, Escherichia coli, Enterococcus faecalis, Acinetobacter baumannii, Staphylococcus aureus, purchased from Beijing Beina Chuanglian Biotechnology Research Institute) as simulated experimental strains, used The determined optimal primer combination and the most suitable primer concentration were used for two-step PCR, and the strains were identified by high-throughput sequencing analysis.
[0103] 2. The first step of PCR amplification
[0104] The primer pair combinations P1 (5 μM)+P5 (5 μM); P3 (5 μM)+P6 (5 μM) were selected, and the sequences of each primer were the same as step 2 in Example 2. Experimental procedure Refer to step 2 in Example 2 to carry out the first step of PCR amplification.
[0105] 3. The first step of PCR product purification
[0106] a. configure 80% ethanol solution;
[0107]b. Add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com