Antibiotic lysobacter spp gene knockout system as well as construction method and application thereof
An antibiotic lysobacteria, gene knockout technology, applied in microorganism-based methods, biochemical equipment and methods, chemical instruments and methods, etc. And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
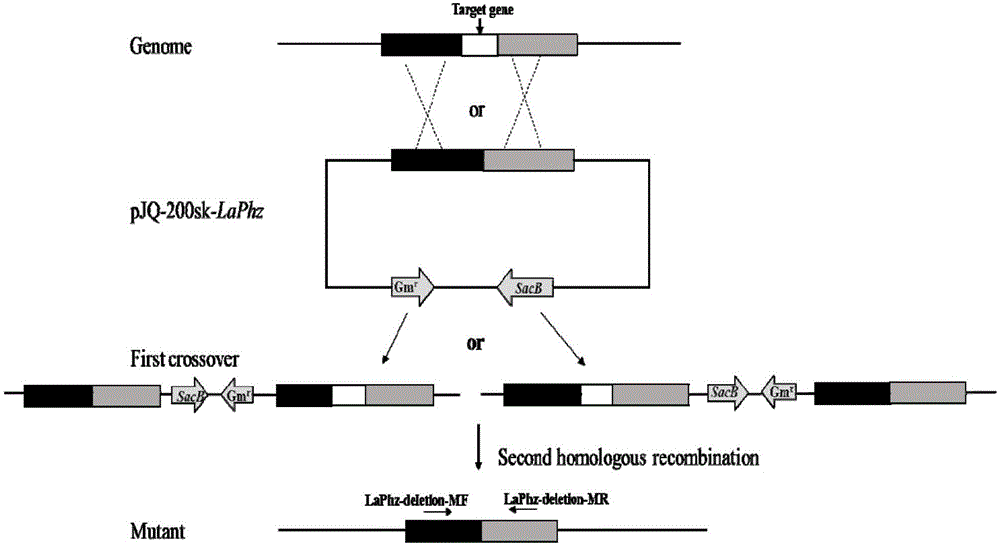
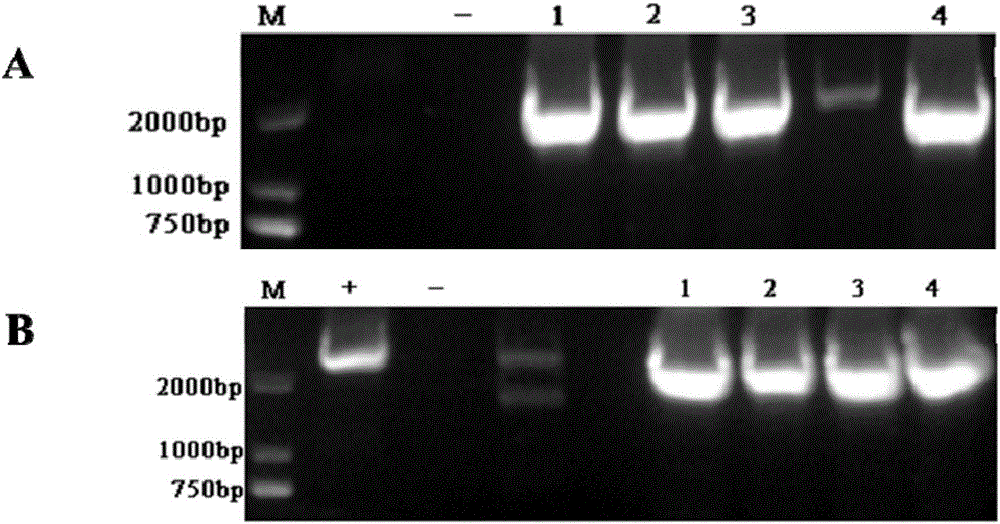
[0043] Knockout of LaPhzD gene in the antibiotic Lysobacterium OH13 of embodiment 1
[0044] Follow these steps:
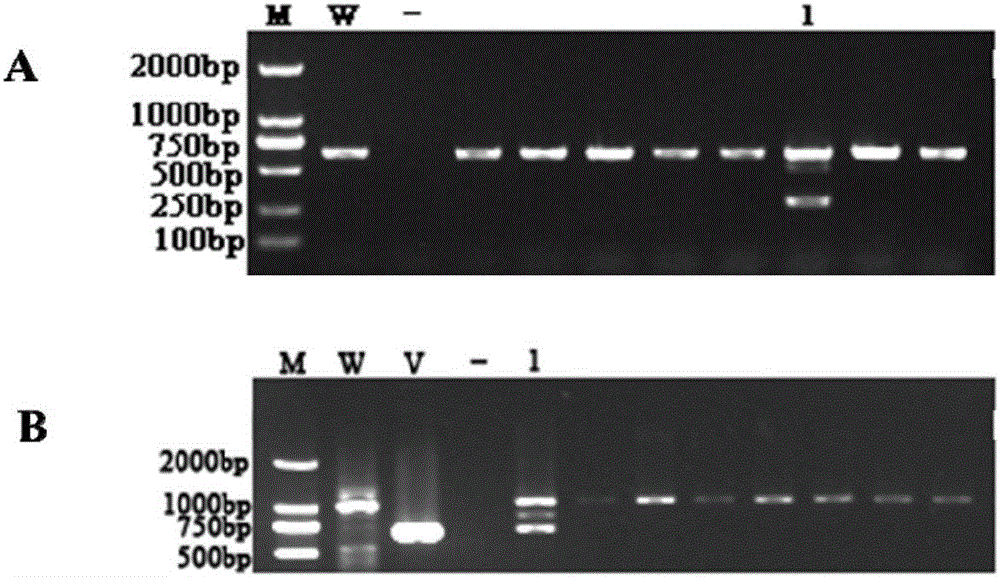
[0045] (1) According to the complete genome information of the antibiotic Lysobacterium OH13, design the target gene LaPhzD upstream arm primer LaPhzD-F1 / LaPhzD-R1 and downstream arm primer LaPhzD-F2 / LaPhzD-R2 and mutant verification primer LaPhzD-deletion-MF / LaPhzD- deletion-MR.
[0046] LaPhzD-F1: CGGGATCCCG ACGAAGCCCTGCTGCTACCC
[0047] LaPhzD-R1: CCCAAGCTTGGG CGAAGCGGCATTCTTGACC
[0048] LaPhzD-F2: CCCAAGCTTGGG ACGCCGTCGCCGATTTCT
[0049] LaPhzD-R2: GCTCTAGAGC CGTGCGCCCTTCGATGAA
[0050] LaPhzD-deletion-MF: AAGATCCAGCCTTATGCG
[0051] LaPhzD-deletion-MR: CAGCAACGGGGTAGTCAT
[0052] (2) The 1100-bp upstream arm was amplified by PCR, and the restriction site was BamHI / HindIII; the downstream arm was 940-bp, and the restriction site was HindIII / XbaI, and the plasmid pJQ200SK was digested with BamHI / XbaI.
[0053] (3) The upper and lower arms after ...
Embodiment 2
[0057] Knockout of LaPhzB gene in the antibiotic Lysobacterium OH13 of embodiment 2
[0058] Follow these steps:
[0059] (1) According to the complete genome information of the antibiotic Lysobacterium OH13, design the target gene LaPhzB upstream arm primer LaPhzB-F1 / LaPhzB-R1 and downstream arm primer LaPhzB-F2 / LaPhzB-R2 and mutant verification primer LaPhzB-deletion-MF / LaPhzB- deletion-MR.
[0060] LaPhzB-F1: CGGGATCCCG CGGAGTCGGACTTGCTGTGG
[0061] LaPhzB-R1: CCCAAGCTTGGG CGCCCTTGCGGCTCATAT
[0062] LaPhzB-F2: CCCAAGCTTGGG GATTCCGGCGATCAAGCG
[0063] LaPhzB-R2: GCTCTAGAGC CAGCAATCCAACCAGGTCTCG
[0064] LaPhzB-deletion-MF: CGGTGAAACCCGACCTGC
[0065] LaPhzB-deletion-MR: GGCGTCTGCCCACTTCTT
[0066] (2) The 1318-bp upstream arm was amplified by PCR, and the restriction site was BamHI / HindIII; the downstream arm was 1102-bp, and the restriction site was HindIII / XbaI, and the plasmid pJQ200SK was digested with BamHI / XbaI.
[0067] (3) The upper and lower arms aft...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com