Kit for detecting esophagus cancer susceptivity and SNP (single nucleotide polymorphism) marker of kit
A technology of susceptibility and markers, applied in the field of genetic testing, can solve problems such as lack of system integration, limitations, and limited detection range, and achieve the effects of good technical reproducibility, high practicability, and high cost performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Used to detect SNP sites associated with susceptibility to esophageal cancer
[0042] Among them, rs10204525 and rs1033667 are located in the region of gene HEATR3, rs1042026 and rs1042522 are located in the region of gene CHEK2, rs10484761 is located in the region of gene ST6GAL1, rs11066015 is located in the region of gene SMG6, rs11066280 is located in the region of gene PTPN2, rs1346044 is located in the region of gene 618ALH1B, 627 is located in the region of gene 618 rs4D27 The gene TMEM173 region, rs1805034 in the gene ATP1B2 region, rs1883965 in the gene HLA region, rs2074356 in the gene PLCE1 region, rs2239612 in the gene MSH2 region, rs2274223 in the gene WRN region, rs238406 in the gene ERCC2 region, rs2847281 in the gene PD-1 region, rs35597309 in the gene region In the ALDH2 region of the gene, rs4785204 is located in the TP53 region of the gene, rs4822983 is located in the RANK region of the gene, rs63749993 is located in the mTOR region of the ge...
Embodiment 2
[0043] Example 2 Kit for detecting susceptibility to esophageal cancer
[0044] 1. Preparation of the kit:
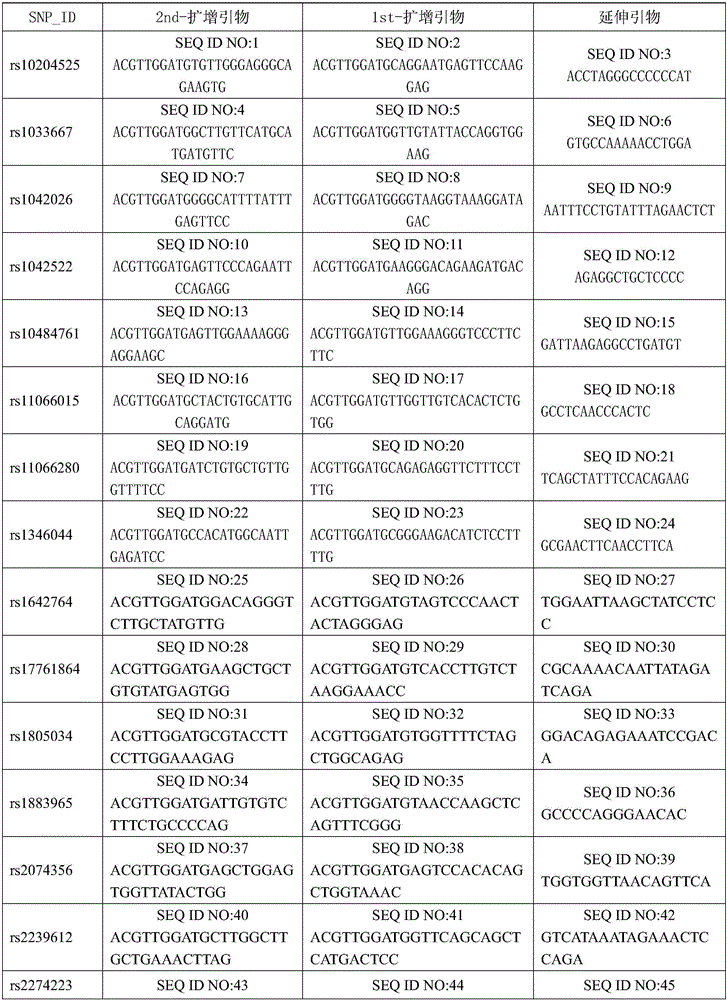
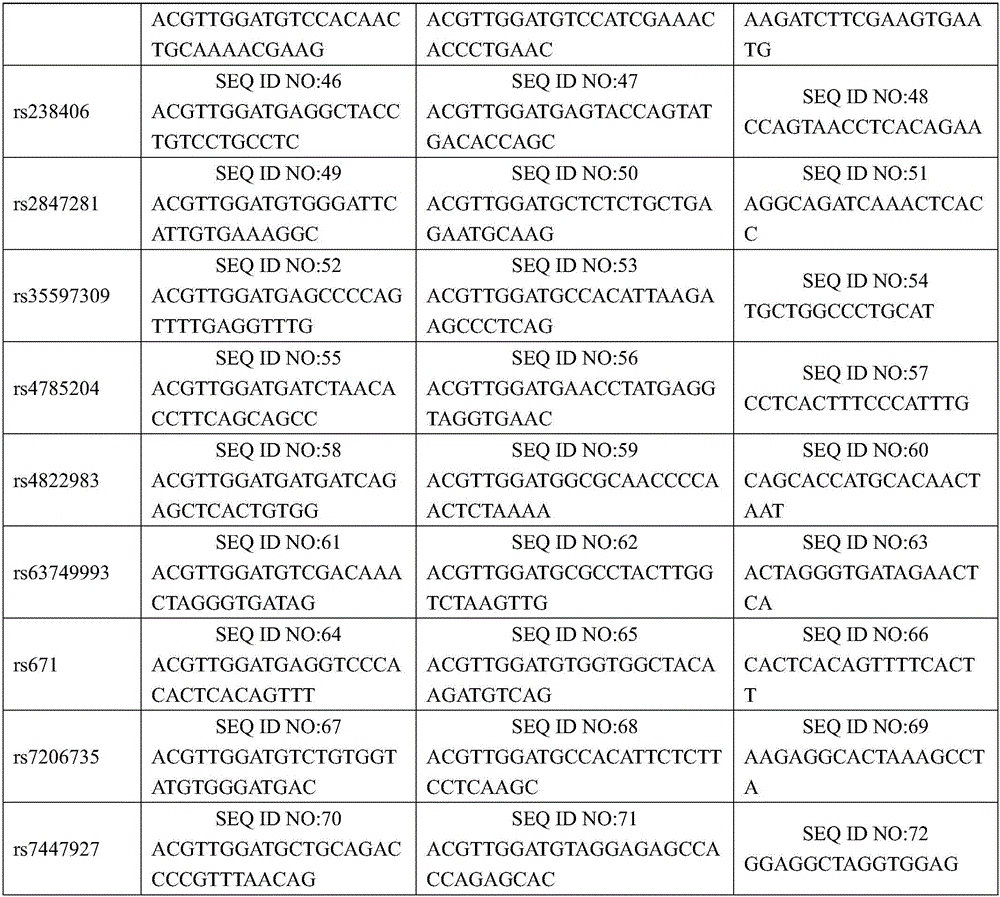
[0045] 1. Design and synthesize PCR amplification primers and single-base extension primers for the 24 SNP sites. The PCR amplification primers and single base extension primers of the SNP sites to be tested are shown in Table 1
[0046] Table 1 PCR amplification primers and single-base extension primers for SNP sites to be tested
[0047]
[0048]
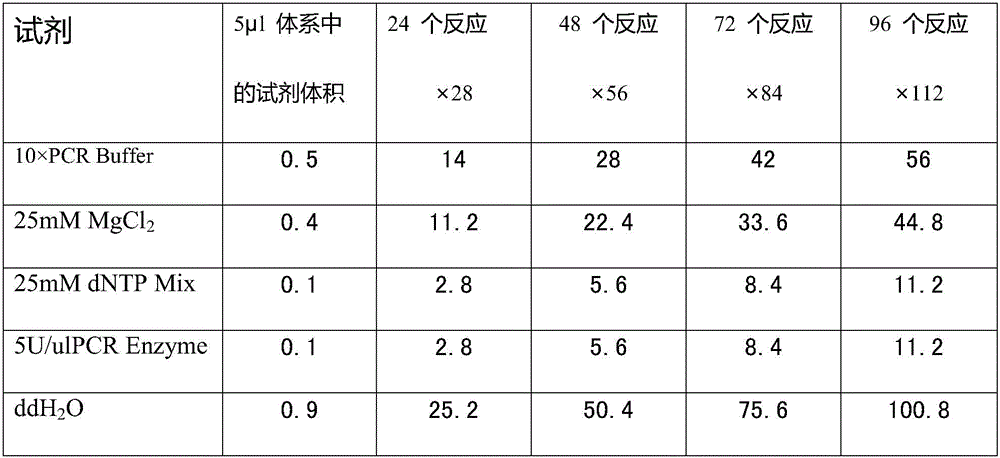
[0049] 2. The kit also includes Taq enzyme, dNTP mixture, MgCl 2 Solution, PCR reaction buffer, enzyme digestion reaction buffer, deionized water.
[0050] 2. The detection method of the kit:
[0051] 1. Extraction of DNA
[0052] 1.1 DNA extraction from oral swab
[0053] 1) Put the oral swab in a 2ml centrifuge tube and add 400μl PBS.
[0054] 2) Add 20 μl QIAGEN Protease and 400 μl Buffer AL. Immediately vortex for 15s to mix. To ensure efficient lysis, sample and Buffer AL must be mixed immediately and th...
Embodiment 3
[0100] Example 3 The literature cited by 24 SNP sites closely related to esophageal cancer, see Table 5
[0101] Table 5 Literature cited by 24 SNP sites closely related to esophageal cancer
[0102]
[0103]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com