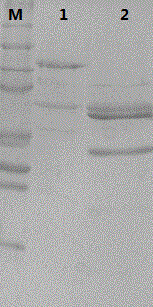
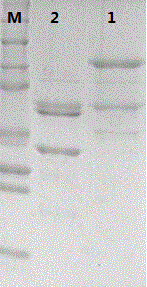
Recombinant human papilloma virus 52 virus-like particle and preparation method thereof
A human papillomavirus, tag-hpv52l1 technology, applied in the field of 52 recombinant human papillomavirus virus-like particles and its preparation, can solve the problems of large protein loss, difficult to apply in large-scale production, and insufficient uniformity of particles
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment l
[0067] Embodiment 1: the design and synthesis of the HPV L1 gene of codon optimization
[0068] The gene sequences were derived from the various types of HPV sequences published on PUBMED. All HPV DNA sequences were synthesized after codon optimization of the selected HPV DNA sequences according to the codon preference of Escherichia coli for gene transcription. Primers were designed according to the synthetic DNA sequence, and the synthetic gene was used as a template for PCR amplification. The resulting codon-optimized sequences were verified by DNA sequencing.
[0069] DNA sequences of various types of HPV before and after optimization:
[0070] SEQ NO.1: DNA sequence of HPV52 type L1 before optimization
[0071] SEQ NO.2: DNA sequence of optimized HPV52 type L1
Embodiment 2
[0072] Example 2: Construction and identification of recombinant vector pGEX-6P-1-GST-HPV52 L1:
[0073] Primers for amplifying the DNA fragment of HPV52 L1: (restriction sites are BamHI and XhoI, respectively)
[0074] Forward-HPV52 L1-ApaI: 5'ACTTCA GGATCC ATGTCTGTTTGGCGTCCGTCTG
[0075] Reverse-HPV52 L1-XhoI: 5'ATCTCA CTCGAG CTA ACGTTTAACTTTTTTTTTTTTT
[0076] PCR Amplification reaction system: 10 x Pfu buffer 20 μL, Pfu enzyme 4 μL, 10 mM dNTP 2.5 μL, 5’ Primer (5 μM) 10 μL, 3’ Primer (5 μM) 10 μL, template DNA 50 ng, add d 2 h 2 0 to 200 μL.
[0077] Gene PCR Amplification conditions: 3 min at 95°C; 30 sec at 95°C, 30 sec at 58°C, 4 min at 72°C; 32 cycles; 10 min at 72°C.
[0078] The L1 gene fragment containing BamH I and XhoI restriction sites and the vector pGEX-6P-1 were subjected to BamH I / XhoI double digestion treatment, and then the recovered gene fragment was combined with pGEX-6P-1 containing the corresponding cohesive end using T4 DNA ligase. ...
Embodiment 3
[0081] Example 3: Construction of recombinant vector pGEX-6P-1m-GST-SUMO-HPV52 L1 vector
[0082] Construction of pGEX-6p-1m vector: In order to make the ApaI restriction site (GGGCCC) near the multi-restriction site the only ApaI restriction site of the vector, a point mutation was carried out without changing the protein expression sequence of the laCl gene ApaI (3890) can be eliminated by changing the Gly codon GGC in another ApaI recognition sequence GGGCCC of the commercially available pGEX-6p-1 vector to its synonymous codon GGT. Through such modification, ApaI can be used to insert and express genes.
[0083] Primers for amplifying the DNA fragment of SUMO: (restriction sites are ApaI and BamHI respectively)
[0084] Forward-SUMO-ApaI: ACTTCA GGGCCC TCTGACCAGGAAGCTAAACCGTC
[0085] Reverse-SUMO-BamHI: CGC GGATCC ACCGGTCTGTTCCTGGTAAAC
[0086] Primers for amplifying the DNA fragment of HPV52 L1: (restriction sites are BamHI and XhoI, respectively)
[0087] Forwar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com