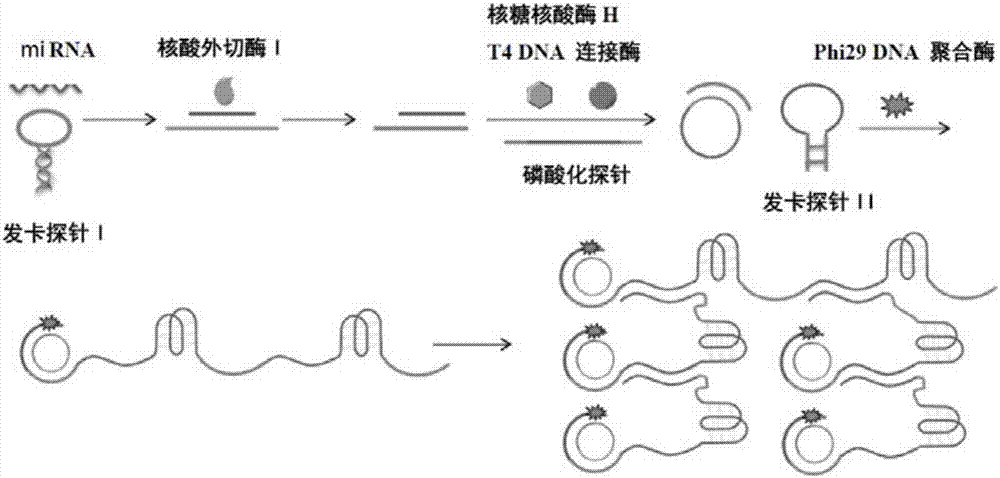
A visualized miRNA detecting method by utilizing an exonuclease reaction to generate primers and dendritic rolling circle amplification
An exonuclease, rolling circle amplification technology, applied in the fields of molecular biology and nucleic acid chemistry, can solve the problems of poor ability to distinguish homologous sequences, large sample volume, complex primer design, etc., to avoid background signal interference, The effect of improving selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] In this example, miRNA21 (sequence 5'-UAGCUUAUCAG ACUGAUGUUGA-3') was used as the detection target.
[0030] 1. Design and synthesis of hairpin probe I, phosphorylated long single-stranded DNA and hairpin probe II
[0031] (1) The I sequence of the hairpin probe is:
[0032] 5'- CCCAACCCATCAACATCAGTCTGATAAGCTA TTACTAG TGGGTTGGG -3'.
[0033] Among them, the underlined sequence can be complementary paired to form a "stem", and the rest form a loop, and the double underlined sequence can be complementary paired with miRNA21 so that the hairpin probe I is opened.
[0034] After hairpin probe I and miRNA21 are complementary paired, they are degraded by exonuclease I and ribonuclease H to obtain a short single-stranded DNA whose sequence is CCCAACCCA TCAACATCAGT CTGATAAGCTA.
[0035] (2) The sequence of the phosphorylated long single-stranded DNA probe is:
[0036] 5'-PO 4 - ACTGATGTTGACAGGAATTAGTAAACAATGAAGACCCAACCCGCCCTACCCTAGCTTATCAG -3'.
[0037] Among them, ...
Embodiment 2
[0058] In this example, miRNA155 is used as the detection target, and the detection method is the same as in Example 1, and the probe sequences used are as follows:
[0059] Hairpin probe I: 5'-CCCAACCCA ACCCCTATCACGATTAGCATTAA TTACTAG TGGGTTGGG-3';
[0060] Phosphorylated long single-stranded DNA probe: 5'-PO 4 - GTGATAGGGGT CAGGAATTAGTAAACAATGAAGACCCAACCCGCCCTACCC TTAATGCTAATC-3';
[0061] Hairpin Probe II: TTAATGCTAATC CAGGAATTAGTAAACAATGAAGA ACCCCTATCAC GATTAGCATTAA.
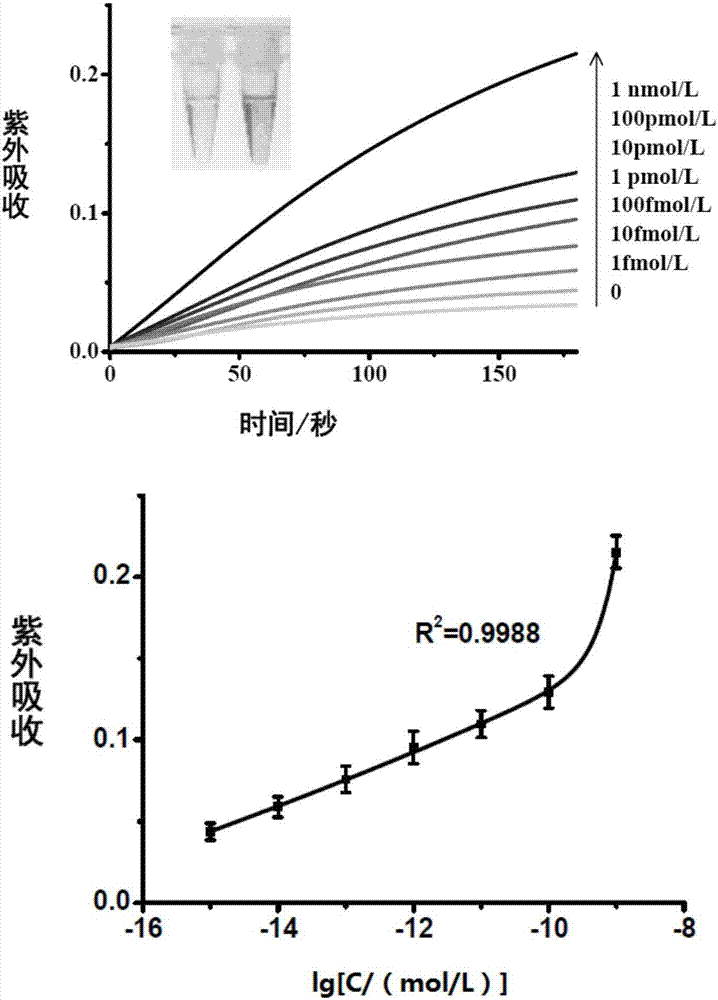
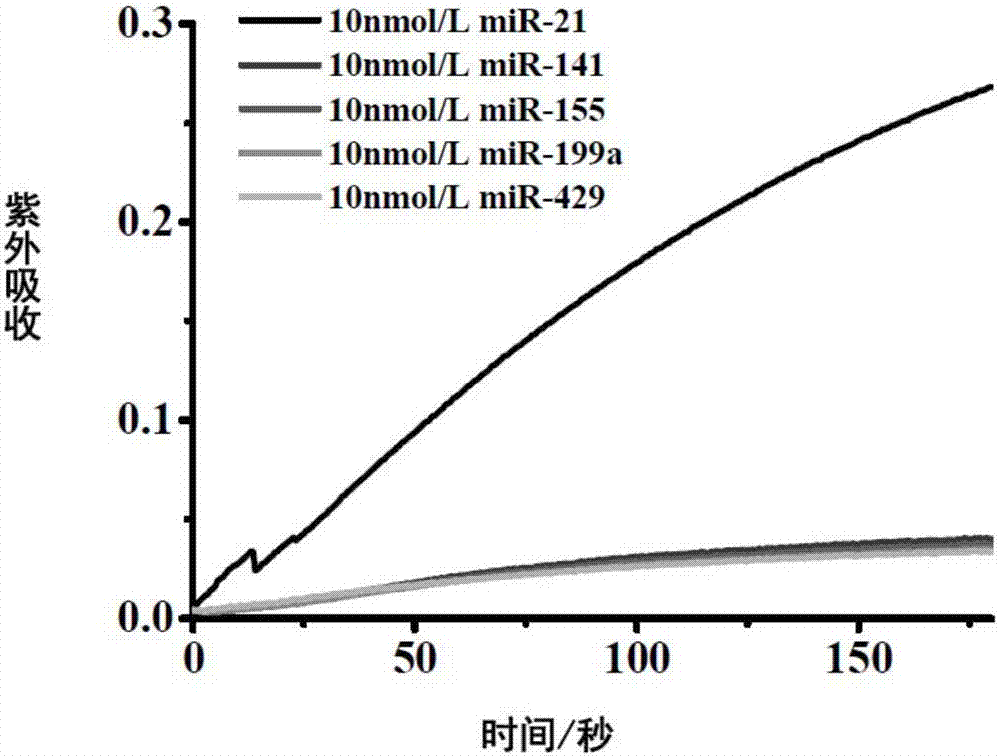
[0062] The result is as Figure 4 , as the concentration of miRNA155 increased, the UV absorption gradually increased. It shows that by changing the probe sequence, the method of the present invention is suitable for detecting other miRNAs.
Embodiment 3
[0064] According to the probe and method in Example 1, the total RNA extracted from the human cervical cancer cell line Hela cells was detected. The amounts of total RNA used were 0.5 μg and 5 μg, respectively. see results Figure 5 , as the amount of total RNA added increases, the UV absorbance value increases.
[0065] The above-mentioned embodiment is a preferred embodiment of the present invention, but the embodiment of the present invention is not limited by the above-mentioned embodiment, and any other changes, modifications, substitutions, combinations, Simplifications should be equivalent replacement methods, and all are included in the protection scope of the present invention.
[0066] SEQUENCE LISTING
[0067] Wuhan University
[0068] A method for visual detection of miRNA using exonuclease reaction to generate primers combined with dendritic rolling circle amplification
[0069] method
[0070] 1
[0071] 6
[0072] PatentIn version 3.3
[0073] 1 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com