Method for detecting polymorphism of folate metabolism related genes through whole-blood direct nucleic acid amplification
A technology for gene polymorphism and nucleic acid amplification, which is applied in the field of primers for direct amplification of nucleic acid in whole blood to detect folic acid metabolism-related gene polymorphisms, can solve the problems of increasing the workload of testing personnel, testing costs, and sample contamination, and achieves Conducive to review, 100% accuracy, simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] PCR of blood samples from different anticoagulant sources: Collect fresh blood from the same person with different anticoagulants, and collect them in EDTA anticoagulant tubes, citrate anticoagulant tubes and heparin anticoagulant tubes respectively.
[0078] Prepare the PCR reaction solution as follows,
[0079] components Volume μl 10×BR Taq Buffer 3 dNTP 2.4 Forward and reverse primer mix 20μM 2 ddH20 19.4 blood sample 3 BR Taq 0.2
[0080] The PCR reaction conditions are as follows:
[0081] 98°C for 3 minutes;
[0082] 98°C for 30s, 58°C for 30s, 72°C for 30s, 40cycle;
[0083] 72℃10min,
[0084] 4℃hold.
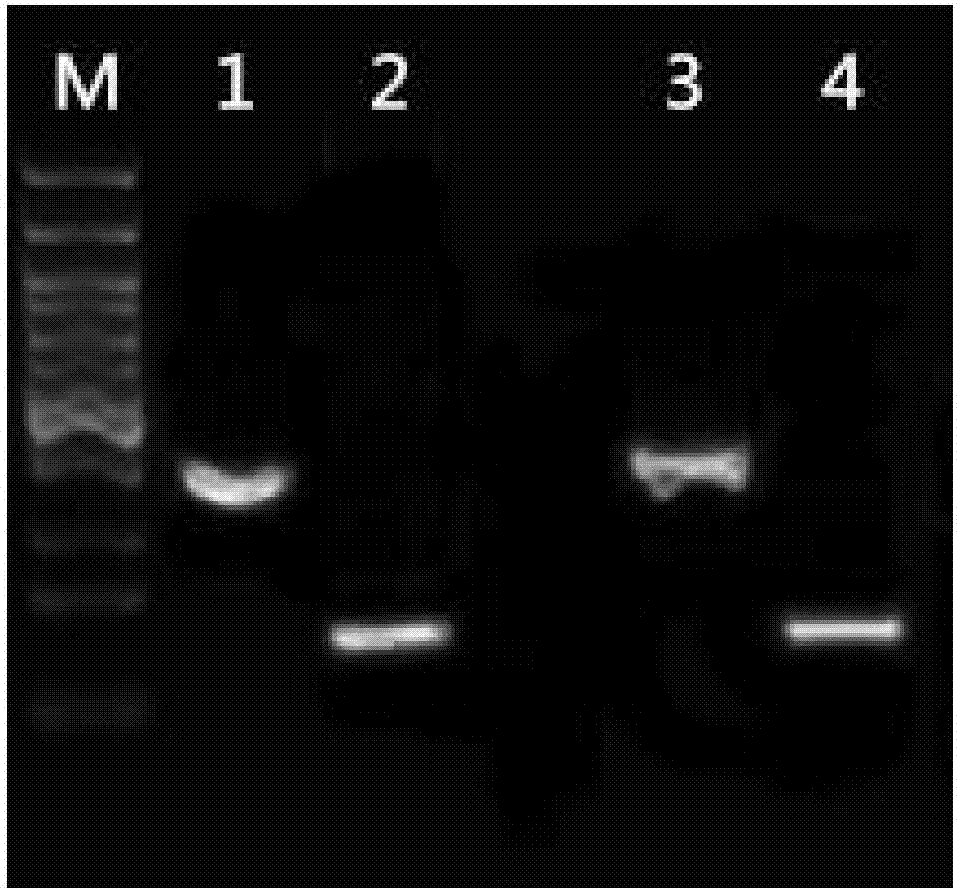
[0085] The PCR electrophoresis images of blood from different anticoagulation sources are shown in figure 2 ,in
[0086] M: 100bp DNA ladder, from top to bottom are 1500bp, 1200bp, 1000bp, 900bp, 800bp, 700bp, 600bp, 500bp, 400bp, 300bp, of which 500bp is a bright band;
[0087] 1: MTHFR-C677T of heparin...
Embodiment 2
[0095] Detect 7 human blood samples from different sources, and prepare the PCR reaction solution according to the following method,
[0096] components Volume μl 10×BR Taq Buffer 3 dNTP 2.4 Forward and reverse primer mix 20μM 2 ddH20 19.4 blood sample 3 BR Taq 0.2
[0097] The PCR reaction conditions are as follows:
[0098] 98°C for 3 minutes;
[0099] 98°C 30s, 58°C 30s, 72°C 30s, 40 cycles;
[0100] 10min at 72°C,
[0101] 4°C hold.
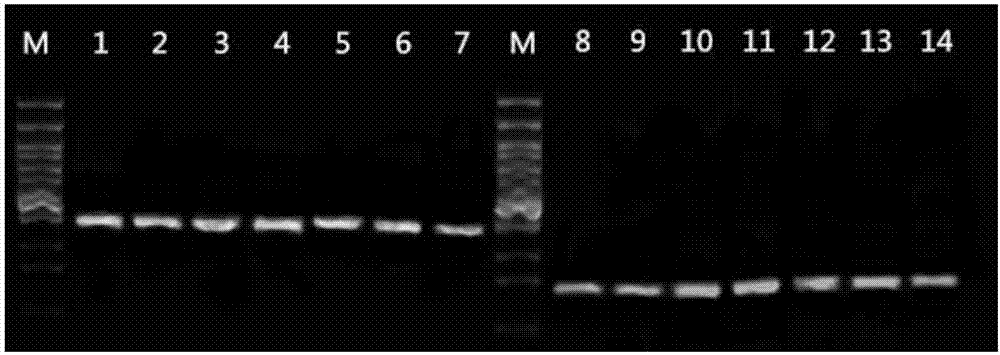
[0102] The PCR electrophoresis graphs of 7 human blood samples from different sources are shown in image 3 ,in
[0103] M: 100bp DNA ladder, from top to bottom are 1500bp, 1200bp, 1000bp, 900bp, 800bp, 700bp, 600bp, 500bp, 400bp, 300bp, 200bp. Among them, 500bp is a bright band.
[0104] 1: MTHFRC677T site of citrate anticoagulated blood sample 1
[0105] 2: MTHFRC677T site of citrate anticoagulated blood sample 2
[0106] 3: MTHFRC677T site of citrate anticoagulated blood sample 3...
Embodiment 3
[0120] The blood of the same sample was tested for 5 times, and the PCR reaction solution was prepared according to the following method,
[0121] components Volume μl 10×BR Taq Buffer 3 dNTP 2.4 Forward and reverse primer mix 20μM 2 ddH20 19.4 blood sample 3 BR Taq 0.2
[0122] The PCR reaction conditions are as follows:
[0123] 98°C for 3 minutes;
[0124] 98°C 30s, 58°C 30s, 72°C 30s, 40cycle;
[0125] 10min at 72°C,
[0126] 4°C hold.
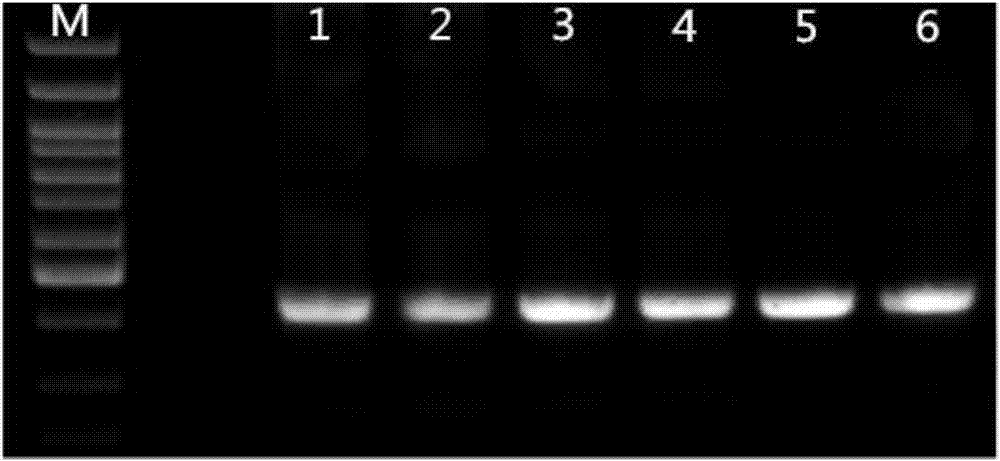
[0127] The PCR electrophoresis figure of the blood of this sample is repeated 5 times to see Figure 4 ,in
[0128] M: 100bp DNA ladder, from top to bottom are 1500bp, 1200bp, 1000bp, 900bp, 800bp, 700bp, 600bp, 500bp, 400bp, 300bp, 200bp. Among them, 500bp is a bright band.
[0129] 1-5: The blood of this sample was tested for MTHFRC677T site repeated 5 times.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com