Method for detecting DNA variation level by using polymolecular tags
A label detection and multi-molecular technology, applied in the field of DNA variation detection, can solve the problems of indistinguishability, data waste, and high cost, and achieve the effect of reducing cost, improving accuracy, and lowering the threshold
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] 1. Synthesis and annealing of the adapter
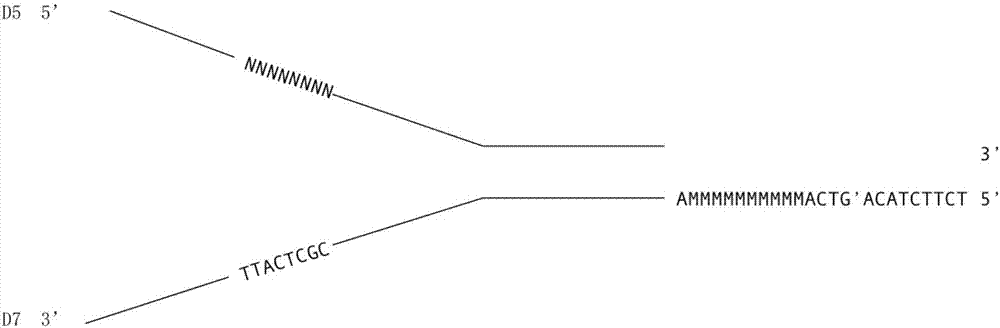
[0040] 1) Synthesize the following sequences in Table 1, wherein NNNNNNNNN in J-AdD5-8N represents the first DNA molecule tag N with a length of 8bp 1 , in J-AdD703-10M MMMMMMMMMM Represents the second DNA molecule label N with a length of 10bp 2 , where the third DNA molecule labels N 3 It is a pre-synthesized short fragment, which is the bold base sequence CGCTCATT in this sequence.
[0041] Table 1
[0042]
[0043] 2) Centrifuge to collect the Adapter powder at the bottom of the tube.
[0044] 3) Open the Adapter tube cap carefully, and be careful not to let the powder float out. Add the corresponding volume of OAB dissolved powder to J-AdD5-8N and J-AdD703-10M respectively, so that the final concentration is 100 μM, and store the stock solution at -20°C for future use. OAB Buffer (oligomeric annealing buffer), the composition is shown in Table 2:
[0045] Table 2
[0046]
[0047] 4) Take 25 μL of J-AdD5-8N...
Embodiment 2
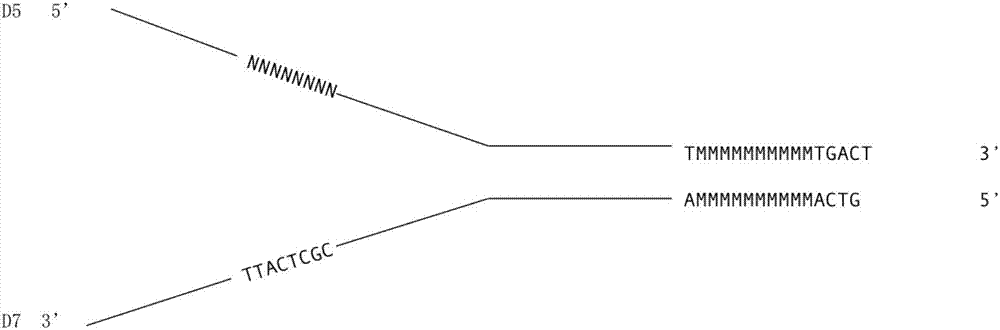
[0076] In the synthesis and annealing reaction of the adapter (adapter) in the first step, the following sequences in Table 7 were synthesized, wherein CGCTCATT in J-AdD5 represents the first DNA molecule tag N with a length of 8bp 1 , in J-AdD703-8N-10N NNN NNN NNNN Represents the second DNA molecule label N with a length of 10bp 2 , NNNNNNNN represents the third DNA molecular label N with a length of 8bp 3 , the synthetic DNA sequence is as follows:
[0077]
[0078] All the other steps are the same as in Example 1.
Embodiment 3
[0080] In the synthesis and annealing reaction of the adapter (adapter) in the first step, the following sequences in Table 8 were synthesized, wherein NNNNNN in J-AdD5-6N represents the first DNA molecule tag N with a length of 6bp 1 , NNNNNN in J-AdD703-6N represents the second DNA molecule label N with a length of 6bp 2 , where the third DNA molecule labels N 3 It is a pre-synthesized short fragment, which is the bold base sequence CGCTCATT in this sequence.
[0081] Table 8
[0082]
[0083] All the following steps are the same as in Example 1. The first DNA molecular label N in each of the above embodiments 1 Sequence, second DNA molecule tag N 2 Sequence and third DNA molecule label N 3 The bases in the sequence are randomly selected from A, T, C and G.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com