Chimeric strong promoter, and applications thereof
A technology of promoters and uses, applied in the field of molecular biology, can solve the problems of weak expression activity, closed expression, and difficulty in meeting the needs of gene therapy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Embodiment 1: Carry the construction of each promoter nucleotide sequence expression vector
[0091] 1. According to the coding sequence of the Renilla luciferase (Renilla luciferase, referred to as RLuc) gene in the psi-CHECK2 plasmid (purchased from Promega Company), a pair of PCR-specific amplification primers (upstream primer F1, plus EcoRI restriction enzyme) were designed at the beginning and end. cutting site and protecting base; downstream primer R1, plus SalI restriction enzyme cutting site and protecting base). The sequence of F1 and R1 is as follows:
[0092] F1: GCCgaattcGCCACCATGACTTCGAAAGT (SEQ ID NO: 5), wherein the lowercase letters represent the EcoRI restriction site
[0093] R1: TGTgtcgacTTATTGTTCATTTTTGAGAACTCGCT (SEQ ID NO: 6), wherein the lowercase letters represent the SalI restriction site.
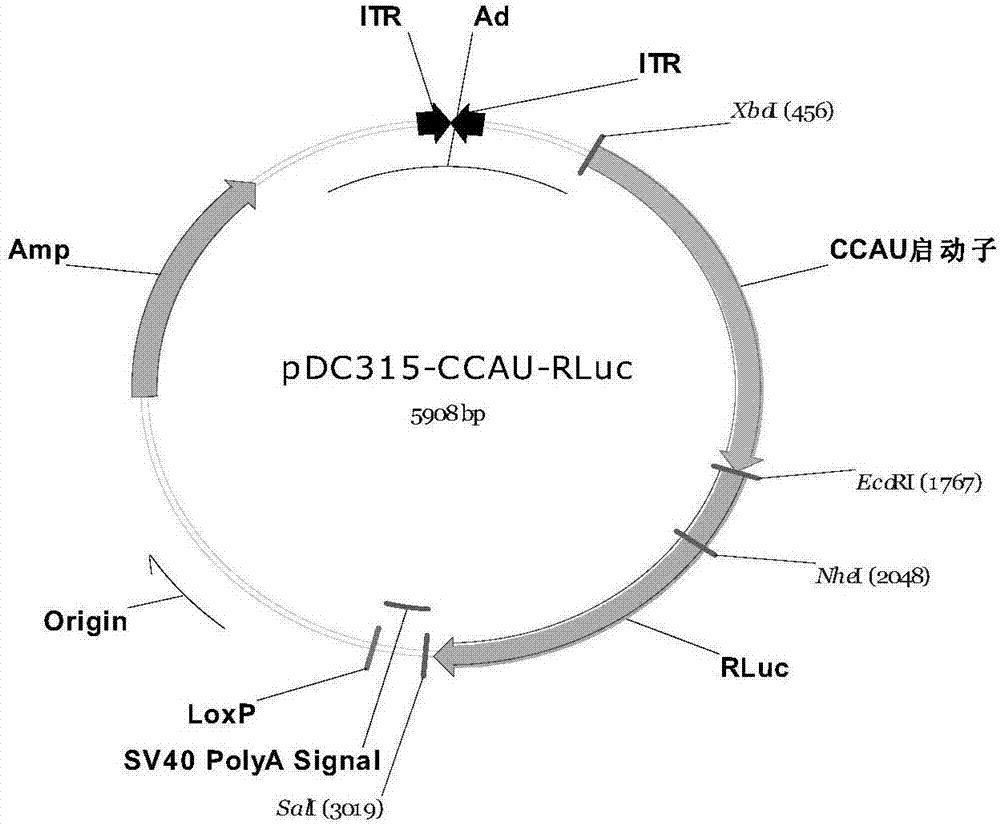
[0094] Using the psi-CHECK2 plasmid as a template, the RLuc gene coding sequence was amplified, digested with EcoRI+SalI, and loaded into the adenovirus...
Embodiment 2
[0101] Embodiment 2: Construction of the adenoviral dual luciferase reporter system carrying each promoter nucleotide sequence
[0102] 1. According to the coding sequence of the firefly luciferase (Fireflyluciferase, referred to as FLuc) gene in the psi-CHECK2 plasmid (purchased from Promega Company), a pair of PCR-specific amplification primers (upstream primer F3, plus EcoRI restriction enzyme) were designed at the beginning and end. cutting site and protecting base; downstream primer R3, plus SalI restriction enzyme cutting site and protecting base). The sequence of F3 and R3 is as follows:
[0103] F3: GCCgaattcGCCACCATGGAAGACGCC (SEQ ID NO: 9), wherein lowercase letters represent the EcoRI restriction site;
[0104] R3: TGTgtcgacTTACACGGCGATCTTTCCGC (SEQ ID NO: 10), wherein the lowercase letters represent the SalI restriction site.
[0105] Using the psi-CHECK2 plasmid as a template, the FLuc gene coding sequence was amplified, digested with EcoRI+SalI, and loaded in...
Embodiment 3
[0109] Example 3: Determination of the activity of each promoter using the adenovirus dual luciferase reporter system
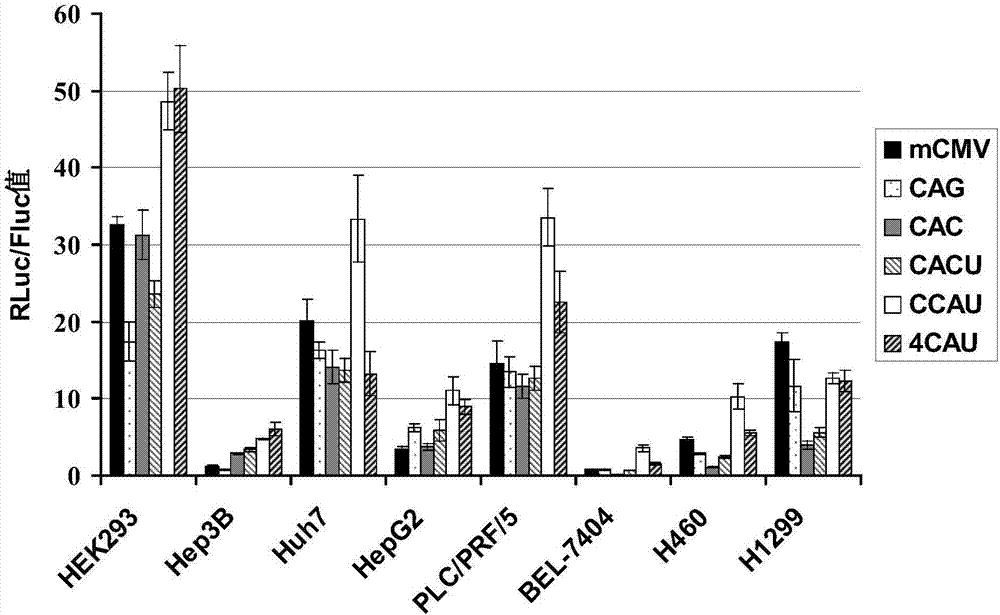
[0110] The low-passage HEK293, Hep3B, Huh7, HepG2, PLC / PRF / 5, BEL-7404, H460, H1299 cell lines (all purchased from ATCC) in good growth state were divided into 1×10 4 Cells / well spread 96-well plate, set at 37°C, 5% CO 2 Culture in the incubator for 24 hours; according to the multiplicity of infection MOI=5, respectively infect Ad-CMV-2Luc, Ad-CAG-2Luc, Ad-CAC-2Luc, Ad-CACU-2Luc, Ad-CCAU-2Luc, Ad-4CAU- 2Luc and other 6 kinds of recombinant viruses were set up with 4 duplicate wells for each group; 2 Incubator culture; after 24 hours, the cells were lysed, and placed in a microplate reader to measure the enzymatic activity of RLuc by a dual-luciferase detection kit (purchased from Promega Company), and the enzymatic activity of FLuc was used as an internal reference to obtain RLuc / FLuc relative ratio. The specific operation steps were completed according ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com