Analytical and diagnostic methods utilizing shigella flexneri apyrase
A technology of Shigella flexneri apyrase and apyrase, applied in biochemical equipment and methods, analytical materials, biological material analysis, etc., can solve the problem of substrate specificity and batch The difference between them cannot achieve complete degradation, influence and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0171] Example 1: Production and colorimetric evaluation of recombinant S. flexneri apyrase (SFA)
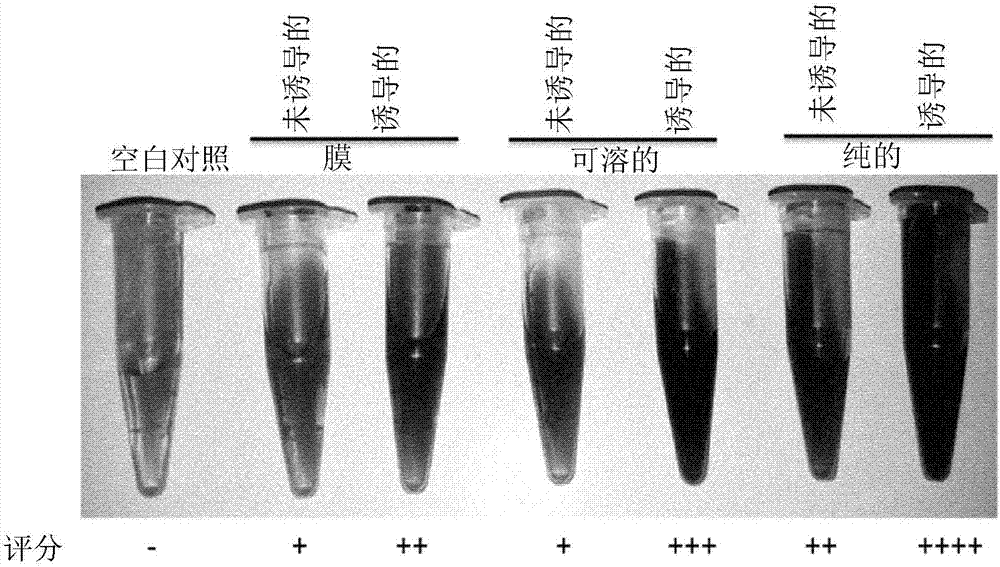
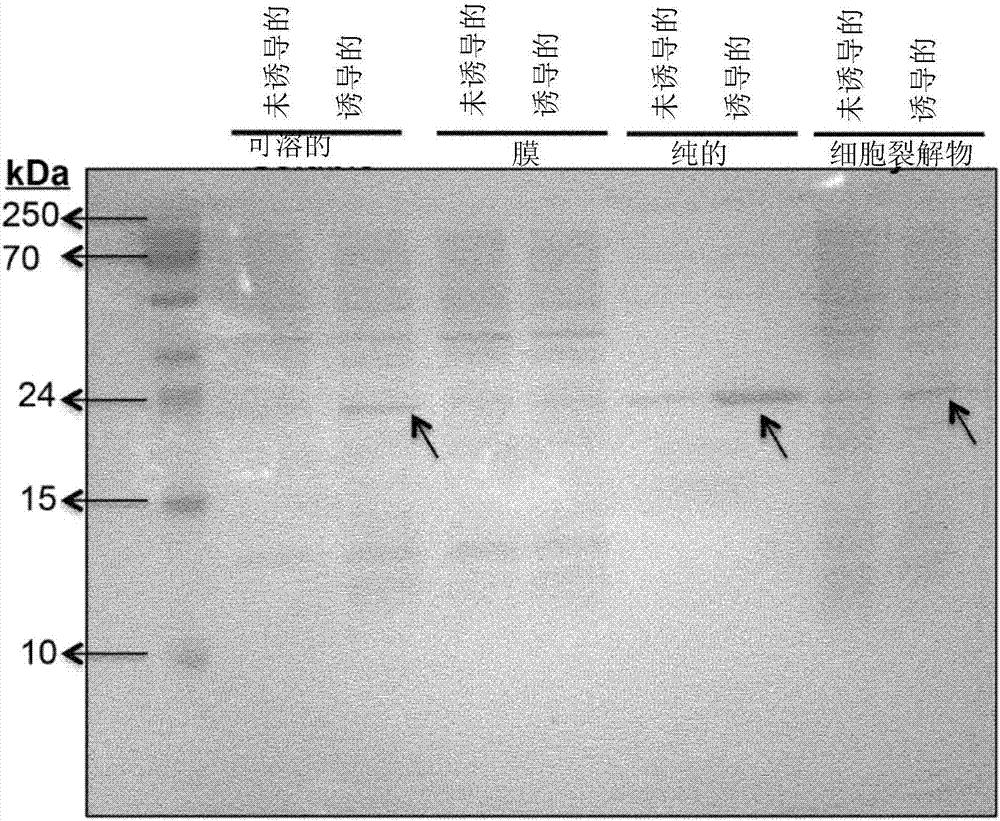
[0172] SFA was prepared and purified as described in the "Materials and Methods" section. Purified enzyme fractions were assayed for the presence of bacterial-derived apyrase using a colorimetric assay. from figure 1 As can be seen, the colorimetric assay clearly shows a color difference that directly corresponds to the expression level of apyrase, revealing its cellular localization. In all cases, a clear color difference between the non-induced and induced cultures could be seen compared to the color of the blank sample.
[0173] from figure 2 As can be seen, there is a clear correlation between the colorimetric analysis results of the clones and the apyrase expression profiles and their localization. The enzyme rSFA was consistently overexpressed at 25 kDa in induced cultures, especially in whole cell lysates and soluble fractions, but not in membrane fractions, thereby ...
Embodiment 2
[0174] Example 2: Sequence analysis between rSFA and STA
[0175] Once protein expression was confirmed by gel electrophoresis and colorimetric analysis, the rSFA gene product was sequenced to determine the nucleotide sequence for comparison with the reference sequence WP_010921592.1 obtained from the NCBI database ( Figure 3A ). As can be seen from the figure, the sequence of the cloned oligonucleotide fragment (rSFA) containing the SFA of apyrase-6xhistidine (Apyrase-6xHis) and the expressed sequence (pBL21-Apyrase-6xHis) fall within the desired range (see Figure 3A ). from Figure 3B As can be seen, the rSFA differs from the reference SFA only at the N- and C-terminus.
example example 3
[0176] Example 3: Compared with STA, rSFA is more efficient and stable in ATP degradation
[0177] Measure the decay of luminescence produced by the firefly luciferase reaction ( Figure 4 ) illustrates the ATP-degrading activity between two apyrases (rSFA and STA). Compared to STA, rSFA exhibited significant removal of ATP and its analogs within ~10 minutes. In other experiments, from Figure 5 It can be seen that the luminescence increases when ATP is added in both cases, whereas the addition of apyrase leads to a decay of luminescence, which is caused by ATP depletion. Deliberate addition of extra ATP during the reaction showed a similar trend of ATP consumption. However, rSFA exhibited faster ATP depletion activity than commercially available STA. When ATP and apyrase preparations were diluted 10-fold, ATP consumption decreased accordingly.
[0178] Stability was checked by storing rSFA samples at 4°C, followed by freezing and freeze-thawing. A study was performed to...
PUM
| Property | Measurement | Unit |
|---|---|---|
| strength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com