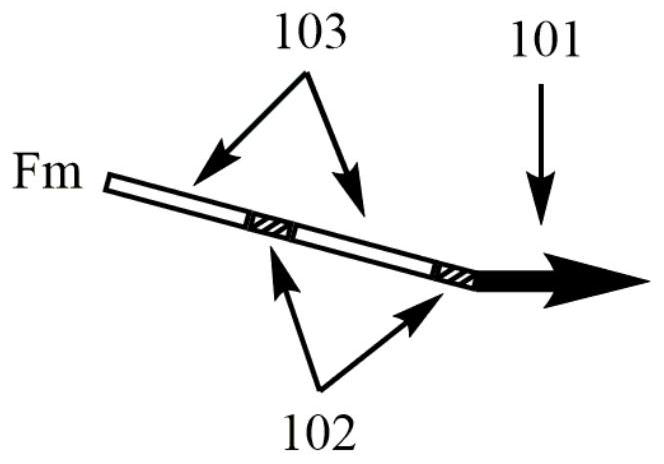
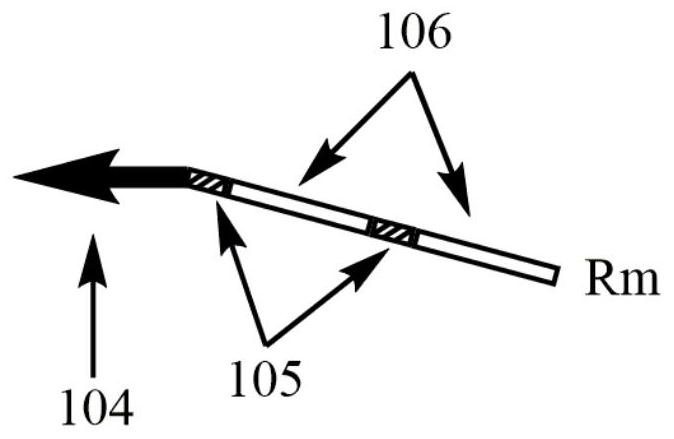
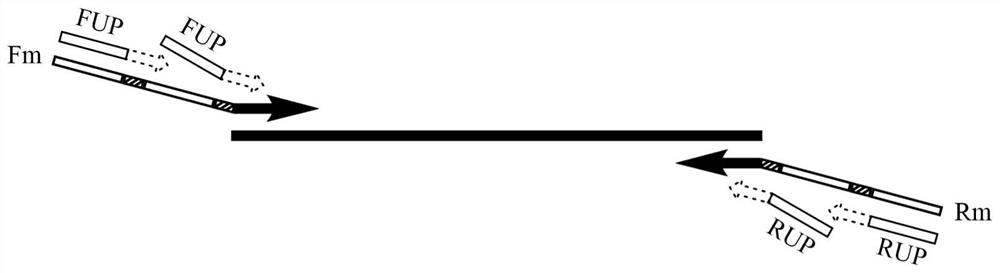A kind of multiple pcr amplification method
An amplified product, one-to-one technology, applied in the field of molecular biology, can solve the problems of limited amplification efficiency and failure to break through the basic mode, and achieve the effects of prolonging the linear accumulation period, high rigor, and improving rigor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Multiplex PCR amplification template DNA:
[0028] Taking the artificially synthesized template of 69 bp as the amplification target, the following primers were designed according to the principle of multiple primer and universal primer design in the technical scheme (Table 1).
[0029] The reaction conditions are as follows:
[0030] The total volume of the PCR reaction system is 25 μL, including 10 U of SD DNA polymerase, 1× SD reaction buffer, and 3.5 mM MgCl 2 , 0.4mM dNTPs, 30nM upstream and downstream multiplex primers Fm and Rm, 1μM upstream and downstream universal primers FUP and RUP, 0.5μL SYBR Geen I (20x), 2μL template. Sterile water was used as negative control.
[0031] The PCR amplification program was: denaturation at 92°C for 2 min, followed by denaturation at 92°C for 20 s, annealing at 56°C for 30 s, extension at 68°C for 20 s, and 25 cycles. Take 5 μL of the above amplification solution and add 1 μL of 6x loading buffer, mix well and take 5 μL for...
Embodiment 2
[0039] Real-time quantitative multiplex PCR detection of Salmonella:
[0040] Multiple primers and universal primers were designed with the ttr gene of Salmonella as the detection target (Table 2).
[0041] The reaction conditions are as follows:
[0042] The total volume of the PCR reaction system is 25 μL, including 10 U of SD DNA polymerase, 1× SD reaction buffer, and 4 mM MgCl 2, 0.4mM dNTPs, 30nM upstream and downstream 2-fold primers F2 and R2 each, 1μM upstream and downstream universal primers FUP and RUP, 0.5μL SYBR Geen I (20x), 2μL template. Sterile water was used as negative control.
[0043] The PCR amplification program was: denaturation at 92°C for 2 min, followed by denaturation at 92°C for 30 s, annealing at 55°C for 30 s, extension at 68°C for 1 min, and fluorescence detection at the extension section, 35 cycles.
[0044] Table 2 List of primer sequences for detection of Salmonella ttr gene
[0045]
[0046] Such as Figure 4 as shown, Figure 4 It is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com