Plant nitrate transporters and uses thereof
一种植物、氨基酸的技术,应用在分子生物学领域,能够解决低氮利用率等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0168] Example 1 - Identification of NRT1.1B / OsNPF6.5
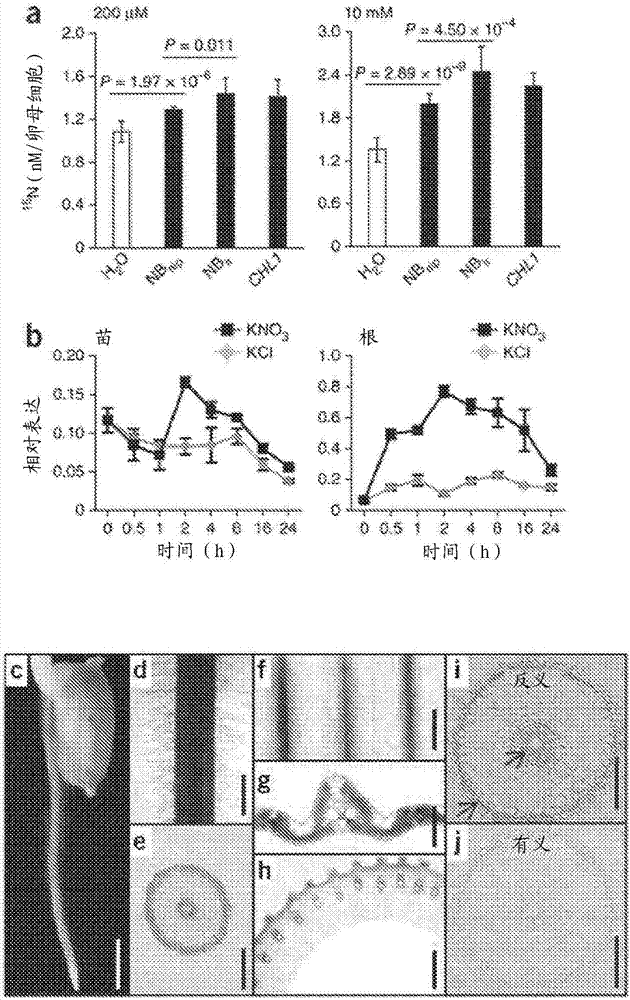
[0169] A wide range of rice varieties for nitrate and ammonium uptake and utilization were analyzed, including 34 indica and japonica cultivars. exist 15 in indica rice after N-nitrate labeling 15 N accumulation was significantly higher than that of japonica rice ( Figure 7 a), while in 15 Between indica and japonica rice after N-ammonium labeling 15 The difference in N accumulation was not statistically significant ( Figure 7 b). This analysis pointed out that indica rice actually had higher nitrate uptake activity than japonica rice. To identify genetic variants associated with divergent use of this nitrate, localization mapping was performed using chlorate, a toxic analog of nitrate. After testing 134 rice cultivars using the chlorate sensitivity assay, indica cultivars could be phenotypically distinguished from japonica cultivars by significantly higher chlorate sensitivity ( Figure 7 c). The...
Embodiment 2
[0179] Embodiment 2——NRT1.1B SNP analysis
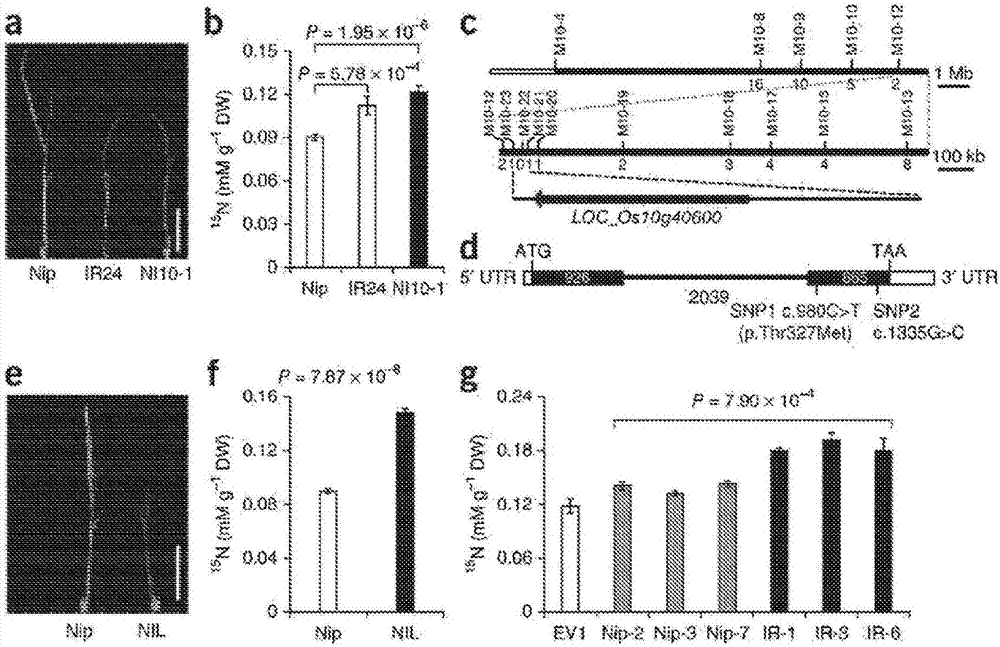
[0180] Sequence analysis revealed two single nucleotide polymorphisms (SNPs) within the coding sequence (CDS) of NRT1.1B between Nipponbare and IR24. SNP1 (c.980C>T) results in a missense mutation where threonine (Thr) in Nipponbare corresponds to methionine (Met) in IR24 (p.Thr327Met substitution), while SNP2 is a synonymous nucleotide substitution (c.1335G>C)( figure 1 d). SNP1 and SNP2 in NRT1.1B were also detected between the parents JX17 and ZYQ8 used for qCHR10 mapping (Table 3), confirming the previous speculation that NRT1.1B corresponds to qCHR10. Amino acid substitutions in NRT1.1B-Nipponbare / IR24 occur in the central cytoplasmic loop (CCL), which is critical for trafficking function. This led us to hypothesize that NRT1.1B mutations caused by SNP1 may be responsible for the divergence in chlorate sensitivity and nitrate utilization between Nipponbare and IR24.
[0181] RNA extraction, cDNA preparation, and...
Embodiment 3
[0187] Example 3 - Verification of the NRT1.1B allele
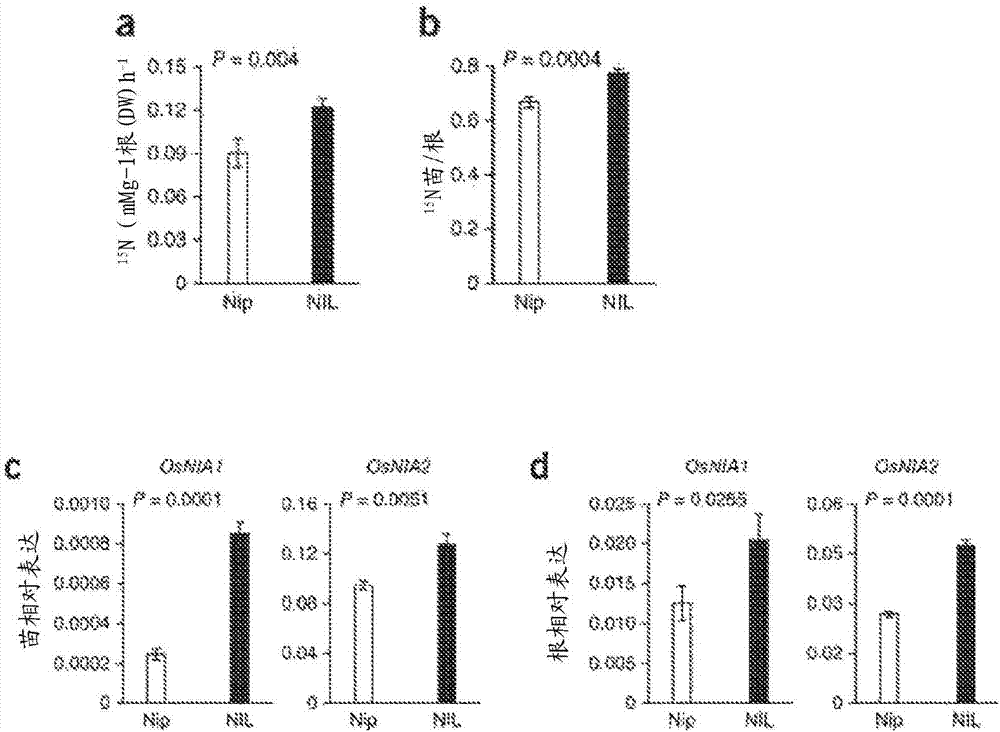
[0188] To test the hypothesis, near isogenic lines (NILs) were further examined, including the NRT1.1B-IR24 allele in the Nipponbare background ( Figure 8 e). Compared with Nippon Fine, NIL is in 15 Chlorate sensitivity was shown after N-nitrate labeling ( figure 1 e) and 15 N accumulation ( figure 1 f and Figure 8 f) significant increase. Transgenic analysis of NRT1.1B-Nipponbare / IR24 under the control of the CaMV 35S promoter or its corresponding native promoter showed that, in 15 15N accumulation in NRT1.1B-IR24 transgenic plants after N-nitrate labeling was higher than in NRT1.1B-Nipponbare transgenic plants ( figure 1 g and Figure 9 a-c). Furthermore, the transcript expression of NRT1.1B in NIL or IR24 was similar or even lower than that of Nipponbare ( Figure 9 d), The possibility that differences in gene expression lead to functional variation in these two NRT1.1B alleles is ruled ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com