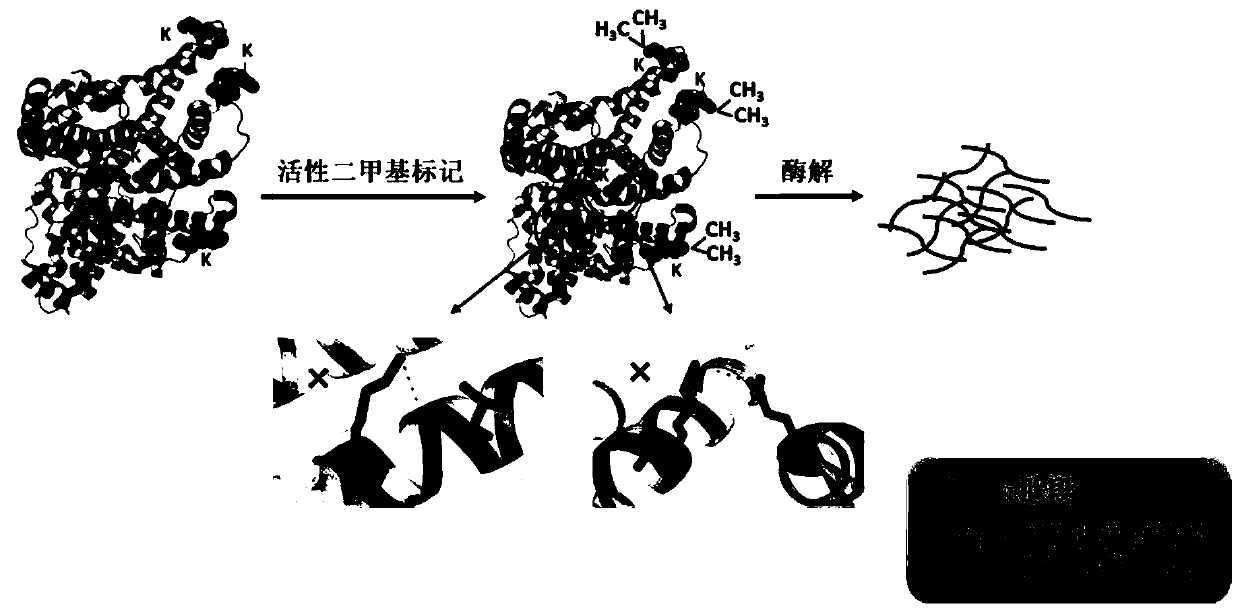
A method for detecting conformational changes of proteins by mass spectrometry
A technology for conformational change, technical detection, applied in the field of active covalent chemical labeling and mass spectrometry detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Active covalent chemical labeling and mass spectrometry detection of protein samples
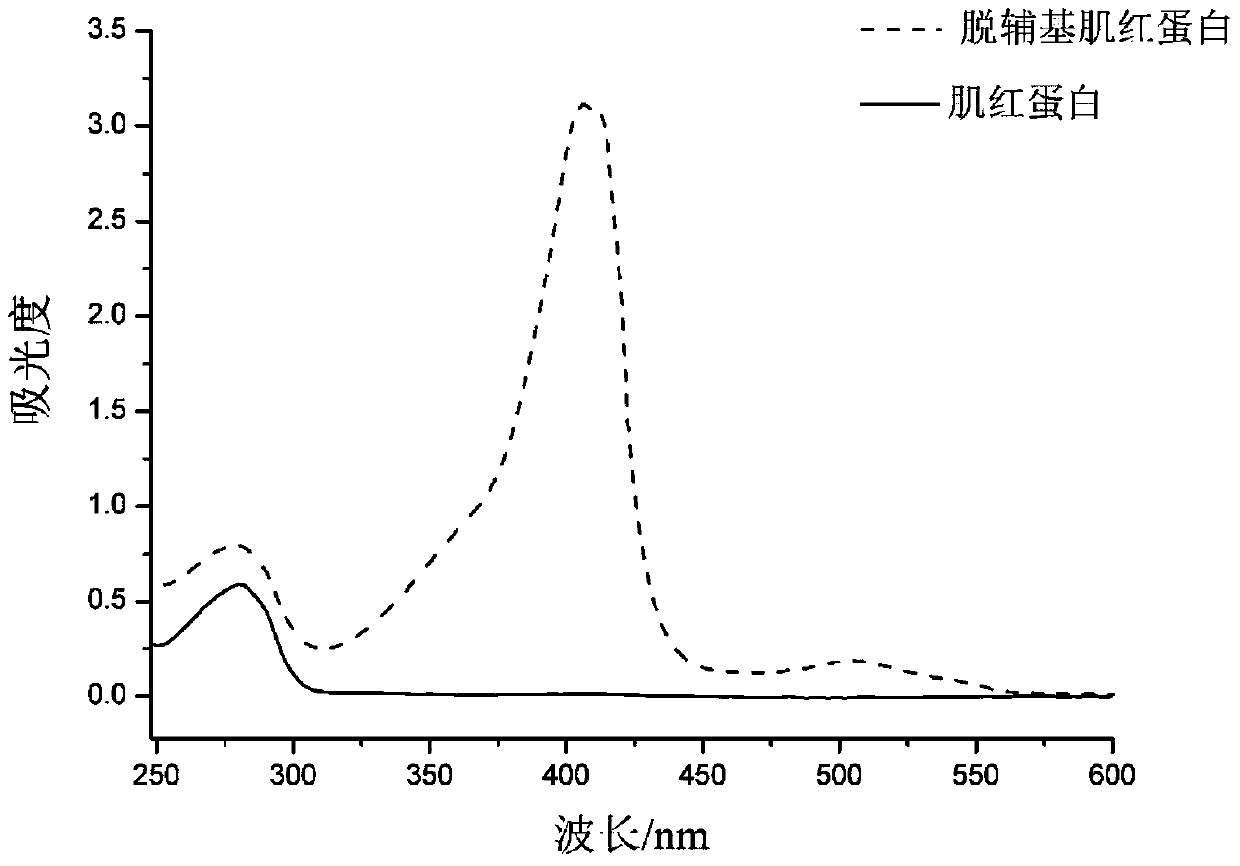
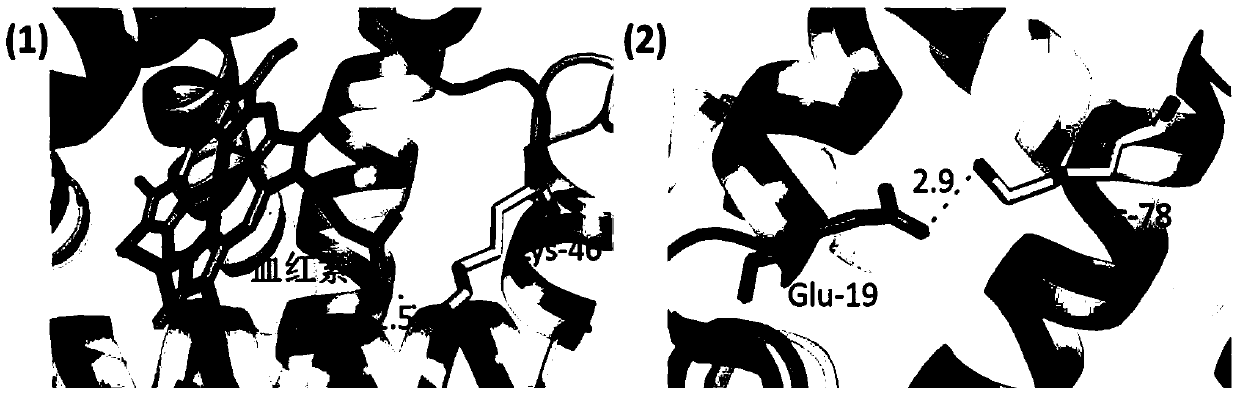
[0049] Myoglobin (Mb) and apomyoglobin (Apomyoglobin, ApoMb) were selected as protein samples. ApoMb is the product of Mb after removing the prosthetic group heme. The structures of the two are very similar, except that the EF, F and FG regions of ApoMb become loose (Eliezer D, Wright P E. Is apomyoglobin a molten globule? Structural characterization by NMR. Journal of molecular biology, 1996, 263(4):531-538).
[0050] (1) Take Mb and ApoMb, each with a mass of 100 μg, and dissolve them in 1 mL of 20 mmol / L HEPES solution at pH 7.0, so that the protein concentration in the solution is 0.1 μg / μL;
[0051] (2) Add 3.2 μL of 4% CH to the protein solution obtained in the above step (1) 2 O and 3.2 μL 0.6mol / L NaBH 3 CN, make NaBH 3 The final concentration of CN was 2mmol / L, and the reaction was carried out at room temperature for 30min;
[0052] (3) Add 50 μL of 1mol / L NH t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com