Method of high-effectively detecting cell cycle of cyanobacteria
A technology for cyanobacteria and Anabaena, which is applied in biochemical equipment and methods, determination/inspection of microorganisms, and introduction of foreign genetic material using vectors, and can solve problems such as the inability of cyanobacteria to detect cell cycle well
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
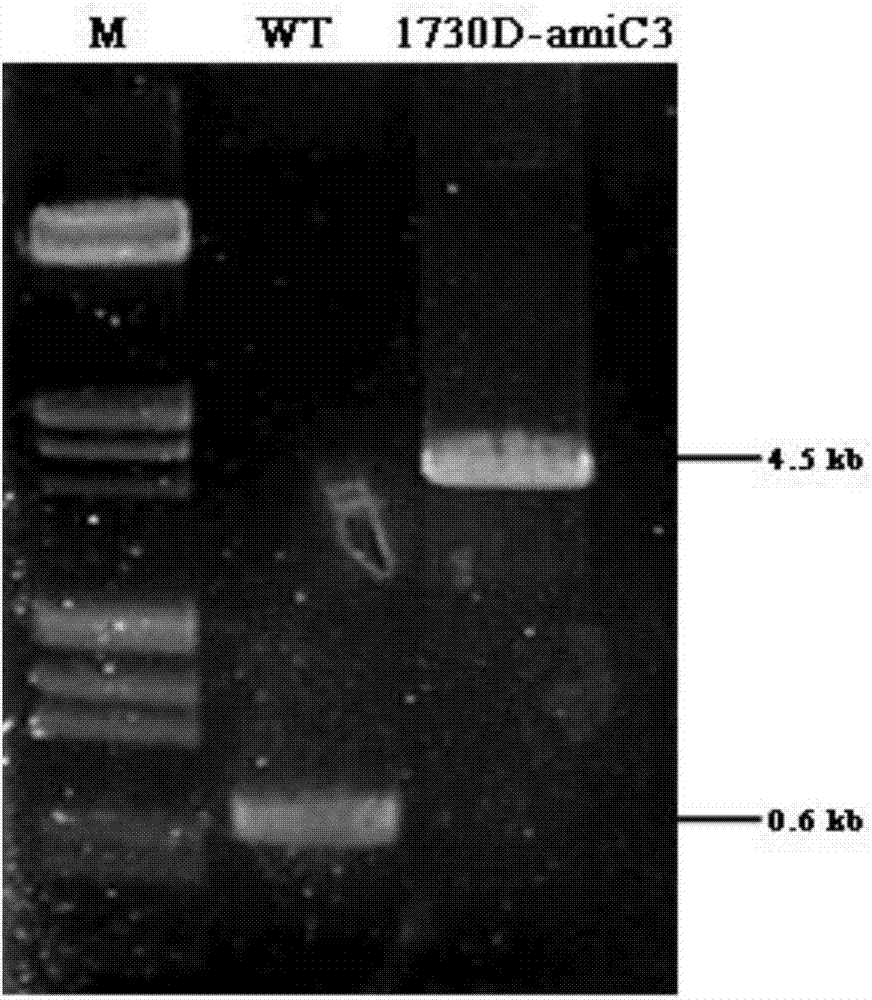
[0026] Embodiment 1, the construction of integration vector and the screening of positive bacterial strain
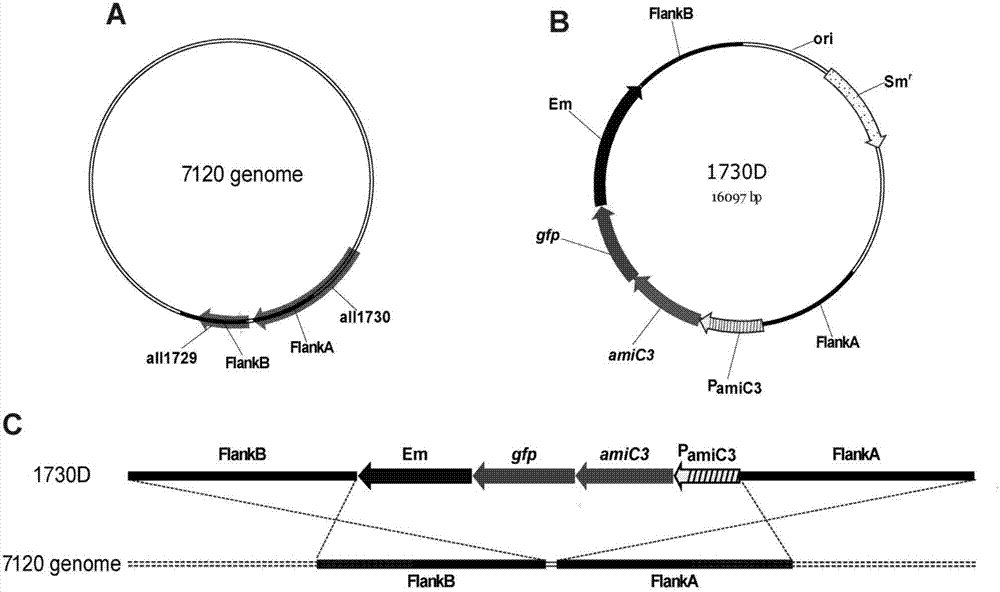
[0027] (1) Construction of integration vector 1730D
[0028] The structure of the integration vector 1730D is as follows figure 1 Shown in middle B, wherein FlankA and FlankB represent homologous fragments on the Anabaena PCC7120 genome respectively (seefigure 1 Middle A). Using the total genomic DNA of wild-type Anabaena sp.PCC 7120 as a template, use primer pairs P1 / P2, P3 / P4 (see Table 1 for primer sequences) to amplify FlanKA and FlanKB fragments, and use primer pairs P5 / P6, P7 / P8 amplifies P amiC3 and amiC3 fragment, use primer pair P9 / P10 to amplify the green fluorescent protein gfp gene fragment, and primer pair P11 / P12 to amplify the erythromycin Em gene fragment; The connection was recovered by enzyme digestion, and the final recovery product was used as a template to amplify the about 7kb fragment of FlanKA-Em-FlanKB with P1 / P4, and then digested with SphI...
Embodiment 2
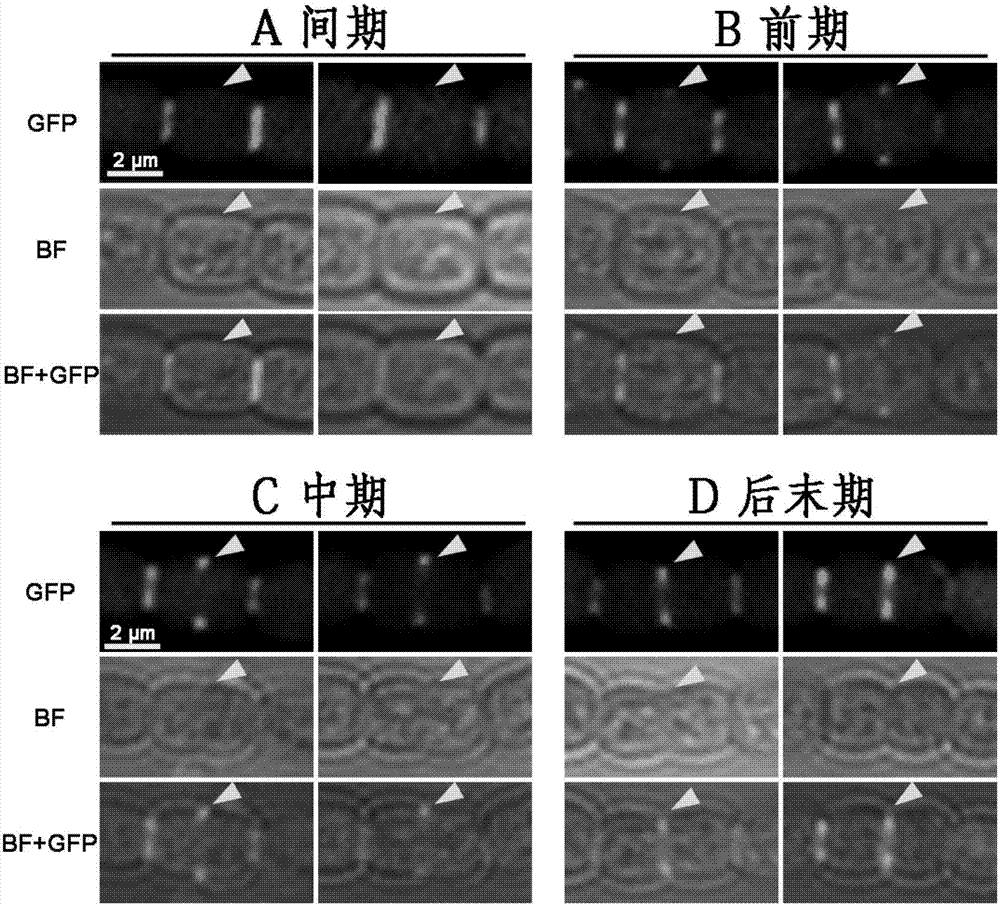
[0040] Embodiment 2, utilize fluorescence microscope to detect the cell cycle of positive bacteria
[0041] After the positive homozygous strain 1730D-amiC3 was cultured in liquid BG11, samples were taken and then observed under a fluorescent microscope. Using the excitation fluorescence at 488nm to observe the green fluorescence of 1730D-amiC3, by image 3 Triangular arrows indicate the result of the cell division ring can be seen: if there is no green fluorescence in the center of the cell, this indicates that the cell is not in the cell division phase, the cell is in interphase ( image 3 Middle A). If there is faint green fluorescence in the center of the cell, this indicates that the cell division ring is assembling and the cell is in prophase ( image 3 Middle B). If there is strong green fluorescence in the center of the cell, this indicates that the cell division ring has been assembled and the cell is in metaphase ( image 3 Middle C). If the green fluorescence i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com