LAMP primer composition for detecting two main parasites causing calf diarrhea and application of LAMP primer composition
A technology of primer combination and calf diarrhea, which is applied in the field of LAMP primer combination of two main parasites, can solve the problems of high primer specificity, high price, long detection cycle, etc., and achieve great promotion value, high sensitivity, highly specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1. Preparation of primer set I and primer set II
[0066] The kit consists of two LAMP primer sets, each of which is used to detect a parasite that causes calf diarrhea.
[0067] The primer set used to detect the parasite Eimeria ITS-2 gene is as follows (5’→3’):
[0068] Outer primer F3 (sequence 1): TTCTACATGTTTGATGCCTTC;
[0069] Outer primer B3 (sequence 2): CCAAACCAAACAATGCTCAA;
[0070] Inner primer FIP (sequence 3): GGAAAAGAAAGGATATGGGCTTGTACGCGGTGTGTCAGAAGT;
[0071] Inner primer BIP (sequence 4): GGTGGATGGATTCTAAGTATTCTCCACACAACACCACCACAGCT;
[0072] Loop primer LF (sequence 5): AGCCATTCATACAACAACAGCAAT;
[0073] Loop primer LB (sequence 6): ATCTCATATTTTAGGAGTTTGGCGA.
[0074] The primer set used to detect the SSU RNA gene of the parasite Cryptosporidium parvum is as follows (5’→3’):
[0075] Outer primer F3 (sequence 7): CGAAAGCATTTGCCAAGG;
[0076] Outer primer B3 (sequence 8): AGGCTCCACTCCTGGTG;
[0077] Inner primer FIP (sequence 9): CAACCTCCAATCTCTAGTTGGCGAACGAAAGT...
Embodiment 2
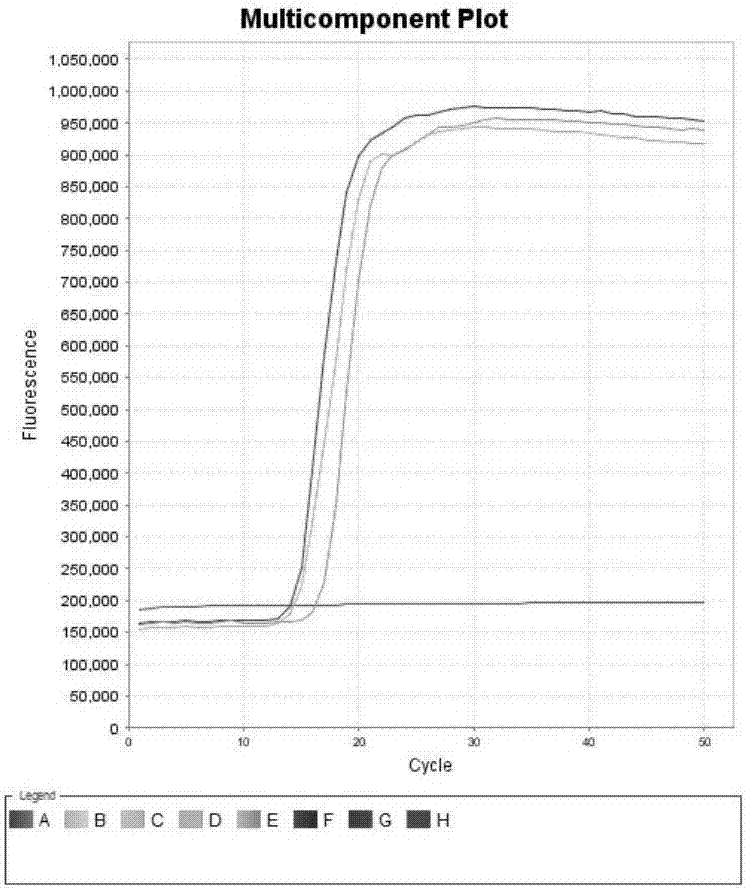
[0085] Example 2. Specificity of primer combination
[0086] 1. Preparation of the sample to be tested
[0087] Sample to be tested 1: Bovine Eimeria ITS-2 gene plasmid.
[0088] Sample to be tested 2: Cryptosporidium parvum SSU RNA gene plasmid.
[0089] The preparation method of each plasmid is as follows:
[0090] Eimeria bovis ITS-2 gene plasmid: Insert the DNA molecule shown in the nucleotide sequence of Genebank No. AB769731.1 between the MCS of plasmid pUC57 (Sangong Bioengineering (Shanghai) Co., Ltd.) to obtain recombination Plasmid, which is the Eimeria bovis ITS-2 gene plasmid.
[0091] Cryptosporidium parvum SSU RNA gene plasmid: Insert a DNA molecule with the nucleotide number of Genebank AF093490.1 between the MCS of plasmid pUC57 (Sanggong Bioengineering (Shanghai) Co., Ltd.) to obtain a recombinant plasmid, which is Resistance gene SSU RNA gene plasmid.
[0092] 2. Testing of samples to be tested
[0093] Each sample to be tested is tested by the following steps:
[0094] ...
Embodiment 3
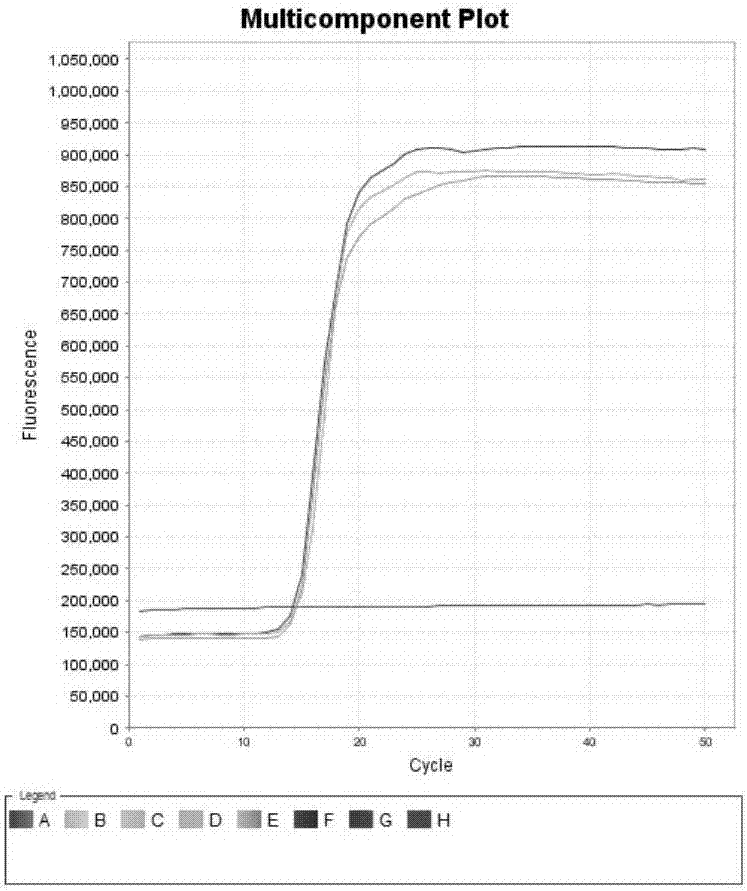
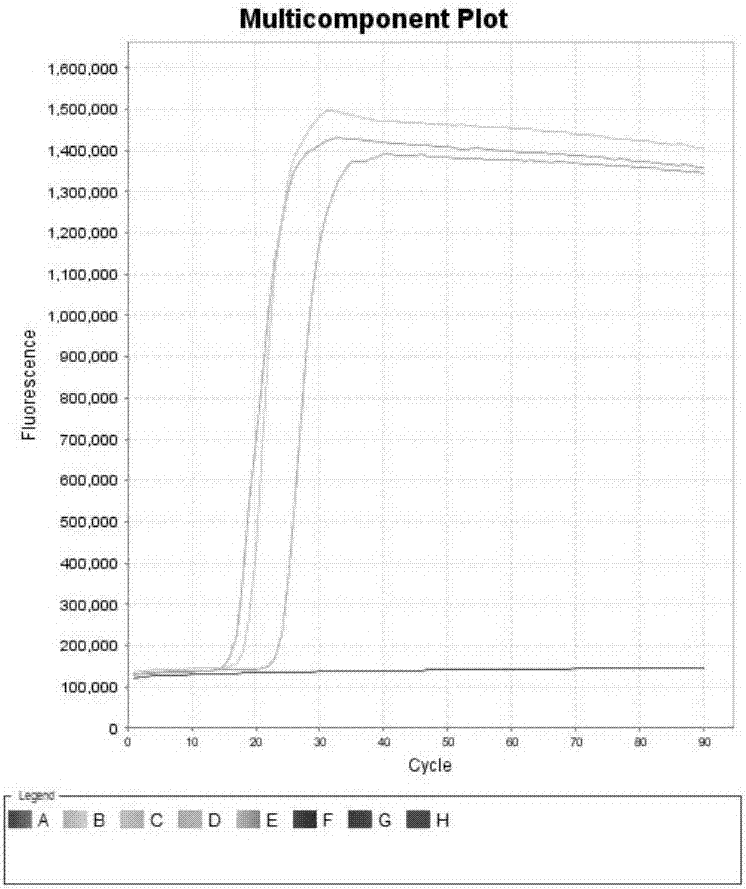
[0101] Example 3. Sensitivity of primer combination
[0102] Sample to be tested 1: The Eimeria bovis ITS-2 gene plasmid of Example 2.
[0103] Sample to be tested 2: Cryptosporidium parvum SSU RNA gene plasmid of Example 2.
[0104] 1. Extract the plasmid DNA of the sample to be tested and dilute it stepwise with sterile water to obtain each dilution.
[0105] 2. Using the diluent obtained in step 1 as a template, the primer set I and primer set II prepared in Example 1 were used for loop-mediated isothermal amplification.
[0106] When the sample to be tested is the sample to be tested 1, use primer set I for loop-mediated isothermal amplification. When the test sample is the test sample 2, the primer set II is used for loop-mediated isothermal amplification.
[0107] Reaction system (10μL): 7.0μL reaction solution (product of Boao Biological Group Co., Ltd., its product catalog number is CP.440020), 1μL primer mixture, 1μL dilution solution (1μL dilution solution contains 10 genome c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com