A highly efficient CRISPR/Cas9 gene editing system optimized for Kluyveromyces
A Kluyveromyces and gene technology, applied in the field of CRISPR/Cas9 high-efficiency gene editing system, can solve the problems of Kluyveromyces industrial production safety hazards, unstable replication and expression, etc., and achieve the effect of safe gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1—KL-CAS1.0 System Transformation
[0036] pKM-Cas9 / gRNA plasmid construction
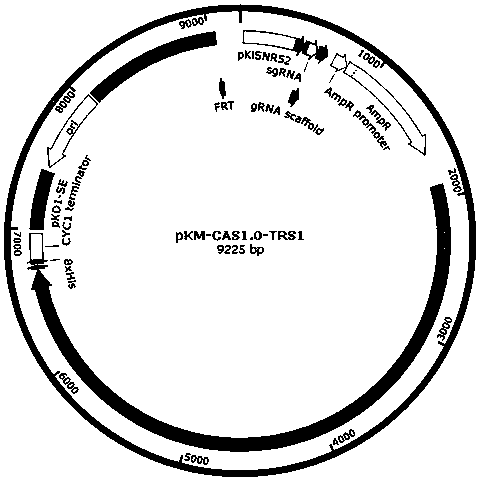
[0037] In the prior art, the pCAS plasmid used in Saccharomyces cerevisiae has both Cas9 gene sequence and gRNA elements, which can realize efficient genome transformation in Saccharomyces cerevisiae through one transformation, but cannot stably replicate and express in Kluyveromyces cerevisiae, such as figure 1 shown. Through transformation, the present invention constructs a new safe and efficient CRISPR / Cas9 gene editing system that is dedicated to Kluyveromyces, can be stably replicated, expressed, and genetically modified in Kluyveromyces, and the gene editing system is named It is KL-CAS1.0. The Kluyveromyces of the present invention is illustrated by taking Kluyveromyces lactis as an example, but not limited thereto.
[0038] Insertion of KL-CAS1.0 efficient pKD1 stabilizing element (SE)
[0039] pKD1 is a commonly used plasmid for Kluyveromyces transformation [13]. In or...
Embodiment 2
[0049] 2. Example 2—Determination of target gene and gRNA sequence
[0050] 2A. Kluyveromyces lactis target gene identification
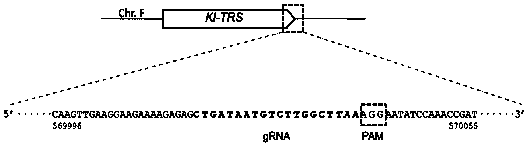
[0051] Sequence search at http: / / www.uniprot.org / , species" Kluyveromyceslactis ",Key words" Threoninet RNA synthetase "" ThrRS "" TRS ". Here, inserting a marker DNA at the tail of this gene is taken as an example. Other target genes or insertion positions and sequences can be operated in a similar manner.
[0052] i. There are two kinds of TRS in K. lactis, which exist in the cytoplasm and mitochondria respectively. After the protein sequence is retrieved, analyze it on the website https: / / ihg.gsf.de / ihg / mitoprot.htmL to determine whether the retrieved protein exists in the cytoplasm (without mitochondrial recognition sequence, eukaryotic homology) or in mitochondria (with mitochondria recognition sequence, prokaryotic homology), pick cytoplasmic TRS Gene (http: / / www.uniprot.org / uniprot / Q6CL41) as the target gene, and named as Kl-TRS ...
Embodiment 3
[0056] 3. Example 3—target sequence pKM-Cas9 plasmid construction
[0057]The present invention replaces the gRNA sequence in the original plasmid with the PCR-homologous recombination method Kl - TRS gRNA sequence. The specific steps are: using the pKM-CAS1.0 plasmid as a template, using primers pKM-Cas9-TRS-F1:CTTTCTGATAATGTCTTGGCTTAAGTTTTAGAGCTAGAAATAGCAAG and primers pKM-Cas9-TRS-R1:GCTCTAAAACTTAAGCCAAGACATTATCAGAAAGTCCCATTCGCCAC for amplification. Mix 17 µL of amplification product, 1 µL of LDpn I, 2 µL of 10×digestion buffer, 37 o C warm bath for 3 h. Add 10 µL of Dpn I-treated product to 100 µL DH5α competent cells, place on ice for 30 min, 42 o After heat shock at C for 45 s, add 1 mL of LB liquid medium37 o C cultured with shaking for 1 h, spread on Amp-resistant LB solid culture, 37 o C was cultured upside down until a single clone grew out. Pick 5 single clones and shake them in LB liquid medium. After the PCR test is positive and confirmed by sequencing, ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com