Detection primers for gene mutation related to spinal muscular atrophy and its application
A spinal muscular atrophy and genetic technology, applied in the detection of spinal muscular atrophy-related gene mutations and genetic testing, can solve the problems of easy errors, high reagent costs, and inability to detect SMN1 gene copy number, etc., and achieve rapid detection results , low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Technical basis
[0025] The invention adopts the fluorescent short-segment multiplex PCR combined with the fragment analysis detection method to detect the mutation of the spinal muscular atrophy related gene. The fluorescent short-segment multiplex PCR method contains two or more pairs of fluorescently labeled primers in one PCR reaction system to detect multiple target sequences. At the same time, the gene analyzer is used to analyze the fragments of the amplified products, and the fluorescent signals of the corresponding fragments can be read, and the quantitative analysis can be carried out by obtaining the peak height and peak area of the fragments.
Embodiment 2
[0026] Example 2: Primer Design
[0027] According to the exon 7 sequence of the SMN1 gene, the primers for specific amplification of the SMN1 gene were designed. Since the SMN1 gene and the SMN2 gene are highly homologous, with only 5 base differences, the 3' end of the upstream primer sequence of the SMN1 gene was specifically amplified. Contains the c.844 C>T difference site located in exon 7, the 3' end of the downstream primer sequence contains the g.27269A>C difference site located in intron 7, and the penultimate base at the 3' end of the downstream primer The mismatch is C, and the specific sequences are SEQ ID NO.1 and SEQ ID NO.2. This primer design can improve the specificity of PCR amplification of SMN1 gene. Three pairs of conserved sequences were selected as internal reference genes, and the specific primer sequences were SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5, SEQ ID NO.6, SEQ ID NO.7, and SEQ ID NO.8. The 5' end of the upstream sequence of all primers was labe...
Embodiment 3
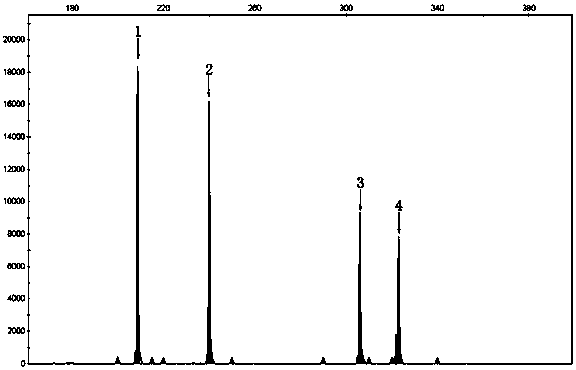
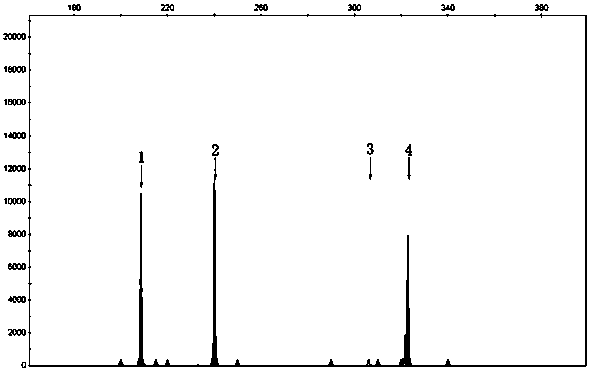
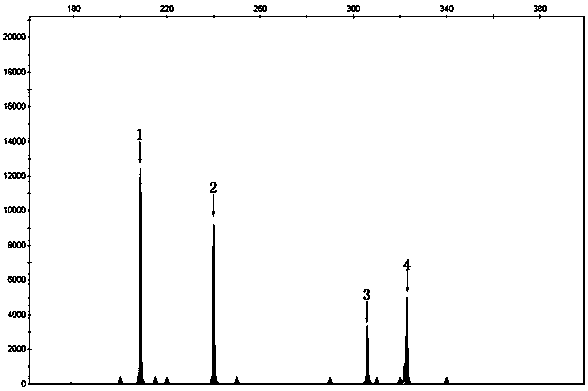
[0028] Example 3: SMN1 Gene Fluorescent Short Fragment Multiplex PCR Detection
[0029]Reaction system: primer mixture 9 μl, 2×Buffer I (Takara, Japan) 12.5 μl, 25Mm MgCl 2 1 μl, 10mM dNTP (Takara Company, Japan) 1 μl, 5U / μl LaTaq enzyme (Takara Company, Japan) 0.25 μl, DNA 50ng.
[0030] PCR instrument: ABI Veriti (ThermoFisher Scientific, USA)
[0031] Reaction conditions: 96°C for 3 minutes; 96°C for 30 seconds, 58°C for 30 seconds, 72°C for 30 seconds, 23 cycles; 72°C for 10 minutes.
[0032] Results: Analysis of 3730 results showed that the fragment size of 307nt was the SMN1 gene, and the fragment size of 209nt, 245nt, and 325nt were the positions of three pairs of internal reference genes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com