Method and kit capable of removing ribosomal RNA with high efficiency for construction of transcriptome sequencing library
A transcriptome sequencing and library technology, applied in the biological field, can solve the problems that affect the detection and analysis of mRNA or LncRNA, fail to effectively remove rRNA kits, rRNA residues, etc., so as to improve the efficiency of sequencing detection and reduce the amount of rRNA residues. , the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
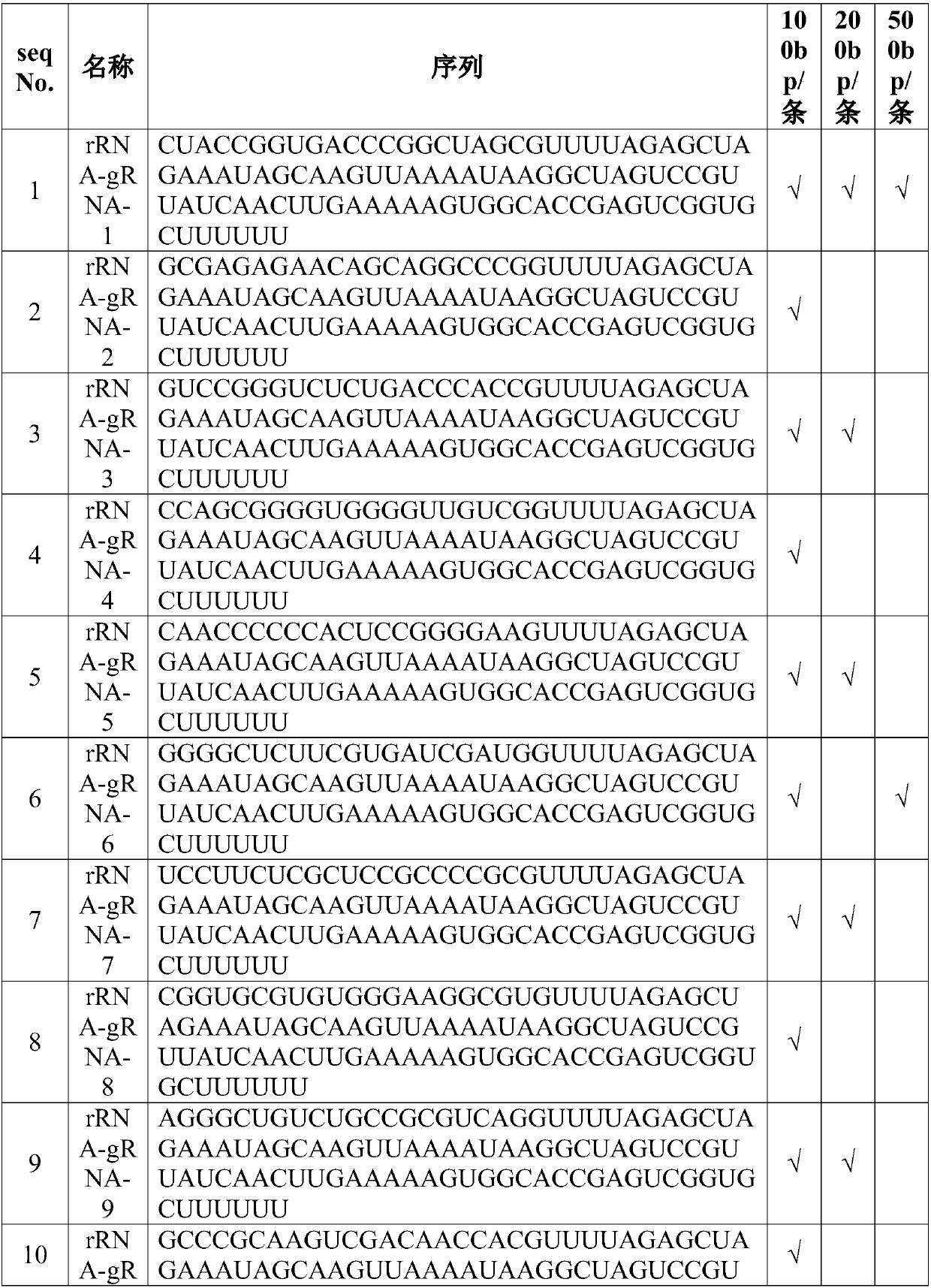
[0036] Example 1: gRNA design density
[0037] 1. Building a database
[0038] 1. Cell culture
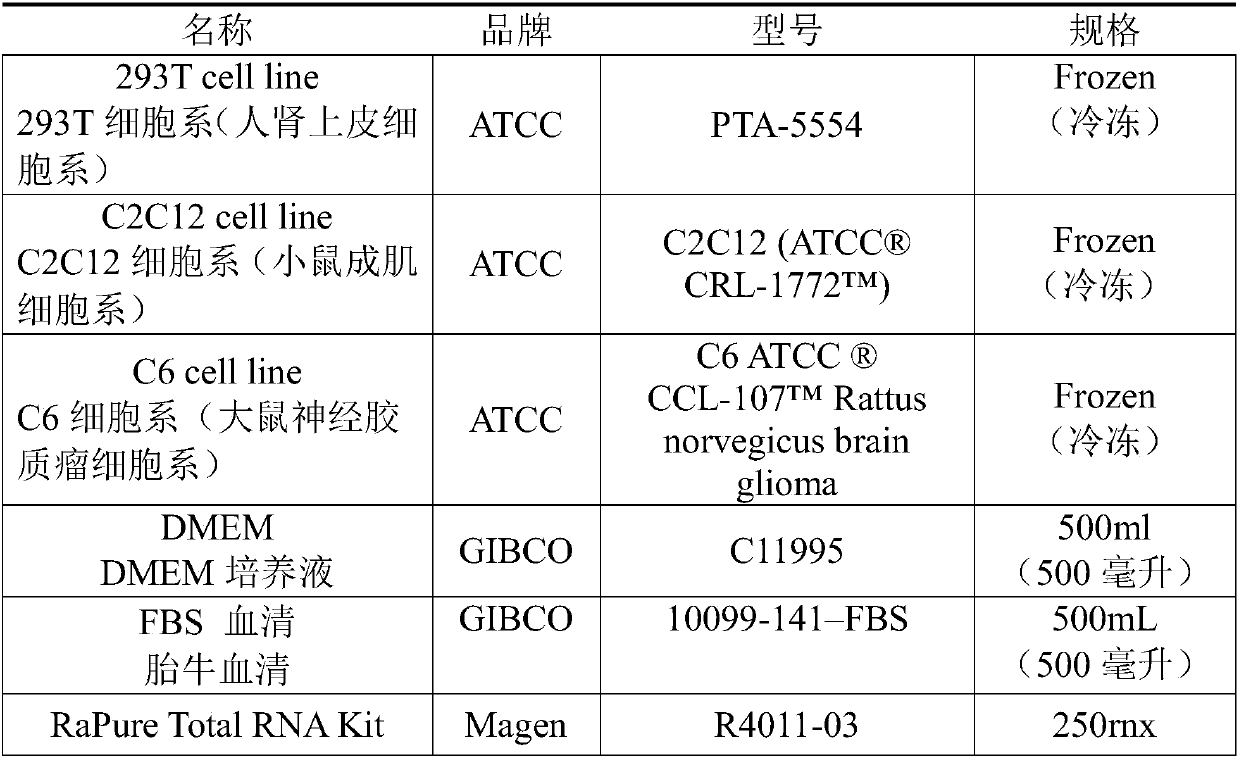
[0039] Use Gibco's DMEM and 10% FBS serum at 37 °C and 5% CO 2 The 293T cells are cultured under culture conditions, and after 2 days of normal culture, the cell density is about 60-80% (40,000-50,000).
[0040] 2. RNA extraction
[0041] Total RNA was extracted according to Magen's RaPure Total RNA Kit instructions.
[0042] 3. RNA library construction:
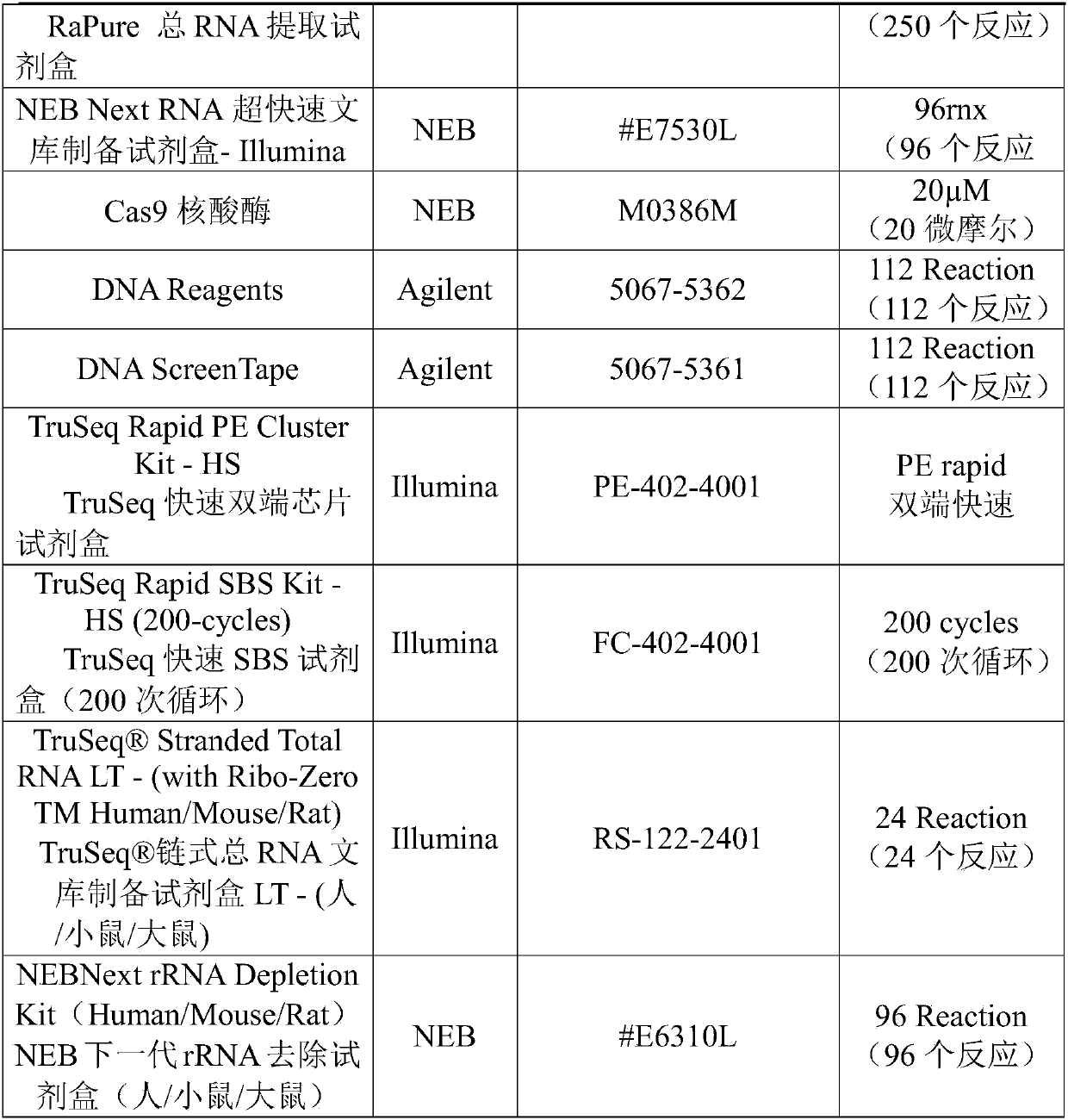
[0043] NEBNext RNA Ultra-Rapid Library Preparation Kit-Illumina was used for library construction with 10ng starting amount (cell RNA), the main steps were: RNA fragmentation (the average fragment length after fragmentation was 200bp); first-strand cDNA synthesis; Second-strand cDNA synthesis and purification; end repair and purification; 3' end addition and purification; adapter ligation and purification; PCR amplification and purification, the cDNA gene library was obtained.
[0044] 2. Library for rRNA depletion
[0045...
Embodiment 2
[0077] Embodiment 2: Cas9 dosage
[0078] 1. RNA library construction
[0079] Concrete method is the same as embodiment one.
[0080] 2. RNA library for rRNA removal
[0081] Concrete method is the same as embodiment one.
[0082] Among them, the design density of gRNApool is 200bp / strip; the working concentration of Cas9 is 30nM, 300nM, 3μM, 30μM and 300μM for experiments.
[0083] 3. On-machine detection of RNA library
[0084] Concrete method is the same as embodiment one.
[0085] 4. Results Analysis
[0086] The results of sequencing analysis showed that the rRNA residues in the rRNA-removed library were all below 30%; finally, the rRNA residues at the dosage of 300nM Cas9 were the least, accounting for only 3.46%, and the number of genes and transcript arrays that could be analyzed were large, respectively. For 26470 and 66887.
[0087]
Embodiment 3
[0088] Embodiment three: reaction time
[0089] 1. RNA library construction
[0090] Concrete method is the same as embodiment one.
[0091] 2. RNA library for rRNA removal
[0092] Concrete method is the same as embodiment one.
[0093] The design density of the gRNA pool is 200bp / strip, the working concentration of Cas9 is 300nM, and the rRNA removal treatment is performed at different times of 5min, 30min, 1h, 2h and 5h respectively.
[0094] 3. On-machine detection of RNA library
[0095] Concrete method is the same as embodiment one.
[0096] 4. Results Analysis
[0097] The results of sequencing analysis showed that the rRNA residues in the library after rRNA removal were all below 20%; when the rRNA removal reaction time was 1 h, the rRNA residues were the least, accounting for only 3.46%, and the number of genes and transcript arrays that could be analyzed were large. 26470 and 66887 respectively.
[0098]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com