A method for predicting spindle assembly checkpoint abnormalities in triple-negative breast cancer using lncRNA-mRNA co-expression network
A technology for triple-negative breast cancer and network prediction, which is applied in the fields of medical data mining, medical informatics, health index calculation, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
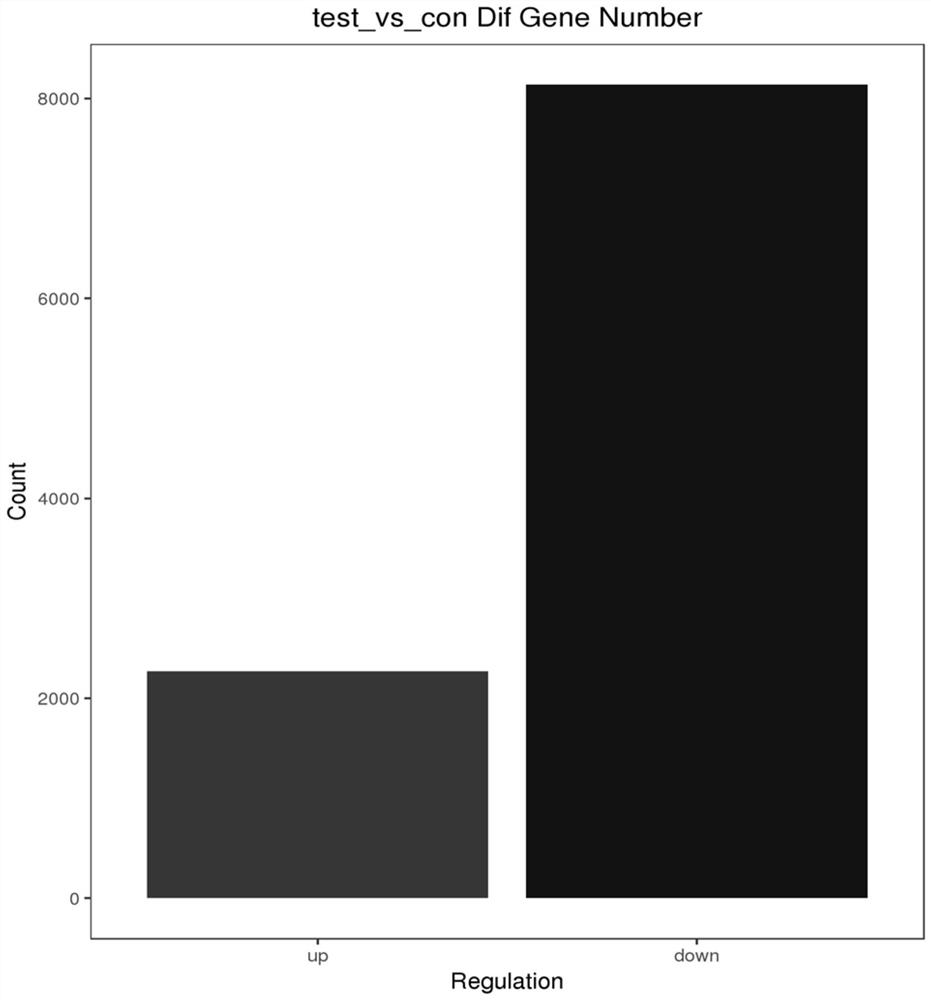
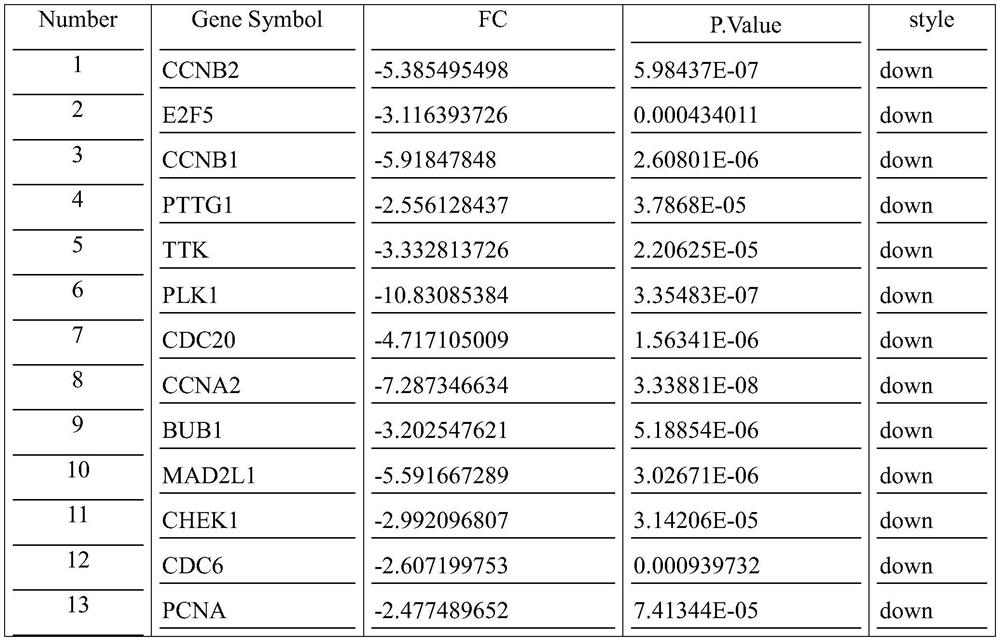
[0040] The method of the present invention using the lncRNA-mRNA co-expression network to predict the abnormal spindle assembly checkpoint of triple-negative breast cancer comprises the following steps:
[0041] Step 1: Experiment Grouping
[0042] Human breast cancer MDA-MB-231 cells were treated with pirarubicin (THP) to establish test group (test group) and con group (comparison group).
[0043] test group: MDA-MB-231 group was treated in 5 μM THP, and each group had 3 biological repetitions;
[0044] con group: MDA-MB-231 group, each with 3 biological replicates.
[0045] Step 2: RNA Extraction
[0046] (1) Remove the culture medium from the cells grown in the logarithmic phase in the 6cm dish, wash twice with PBS and discard.
[0047] (2) Add 1-2 mL of Trizol reagent to the plate, blow and beat the cell layer repeatedly with a pipette and collect it into a centrifuge tube.
[0048] (3) Add 200-500 μL of chloroform into the centrifuge tube, shake it up and down fully u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com