Method and device for processing repetitive sequences of circulating tumor deoxyribonucleic acid (DNA)
A DNA repetition and sequence technology, applied in the field of genetic engineering, can solve the problem of low accuracy, achieve the effect of improving processing accuracy and reducing manual errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
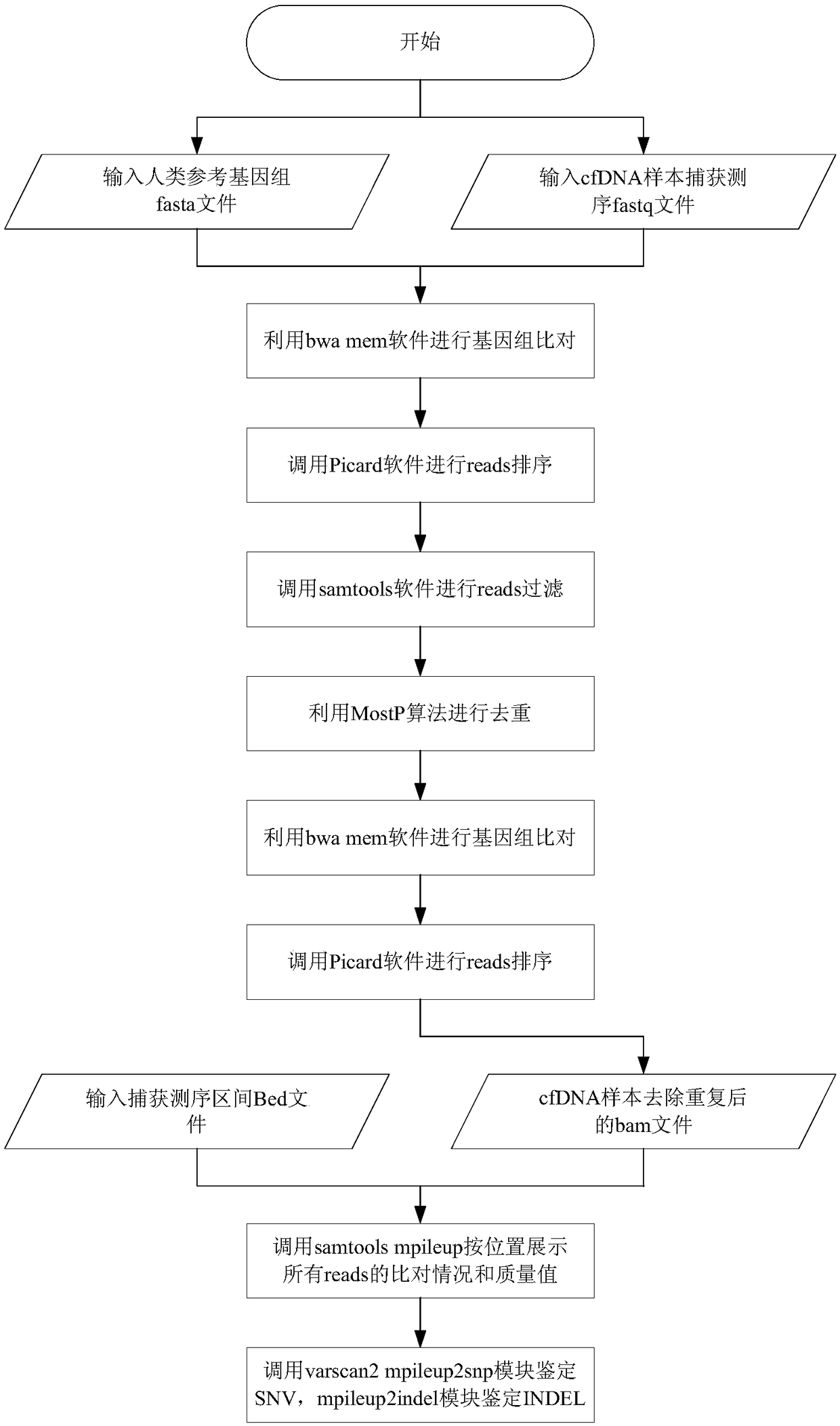
[0043] According to an embodiment of the present invention, an embodiment of a method for processing circulating tumor DNA repeat sequences is provided. It should be noted that the steps shown in the flowcharts of the accompanying drawings can be implemented in a computer system such as a set of computer-executable instructions and, although a logical order is shown in the flowcharts, in some cases the steps shown or described may be performed in an order different from that shown or described herein.
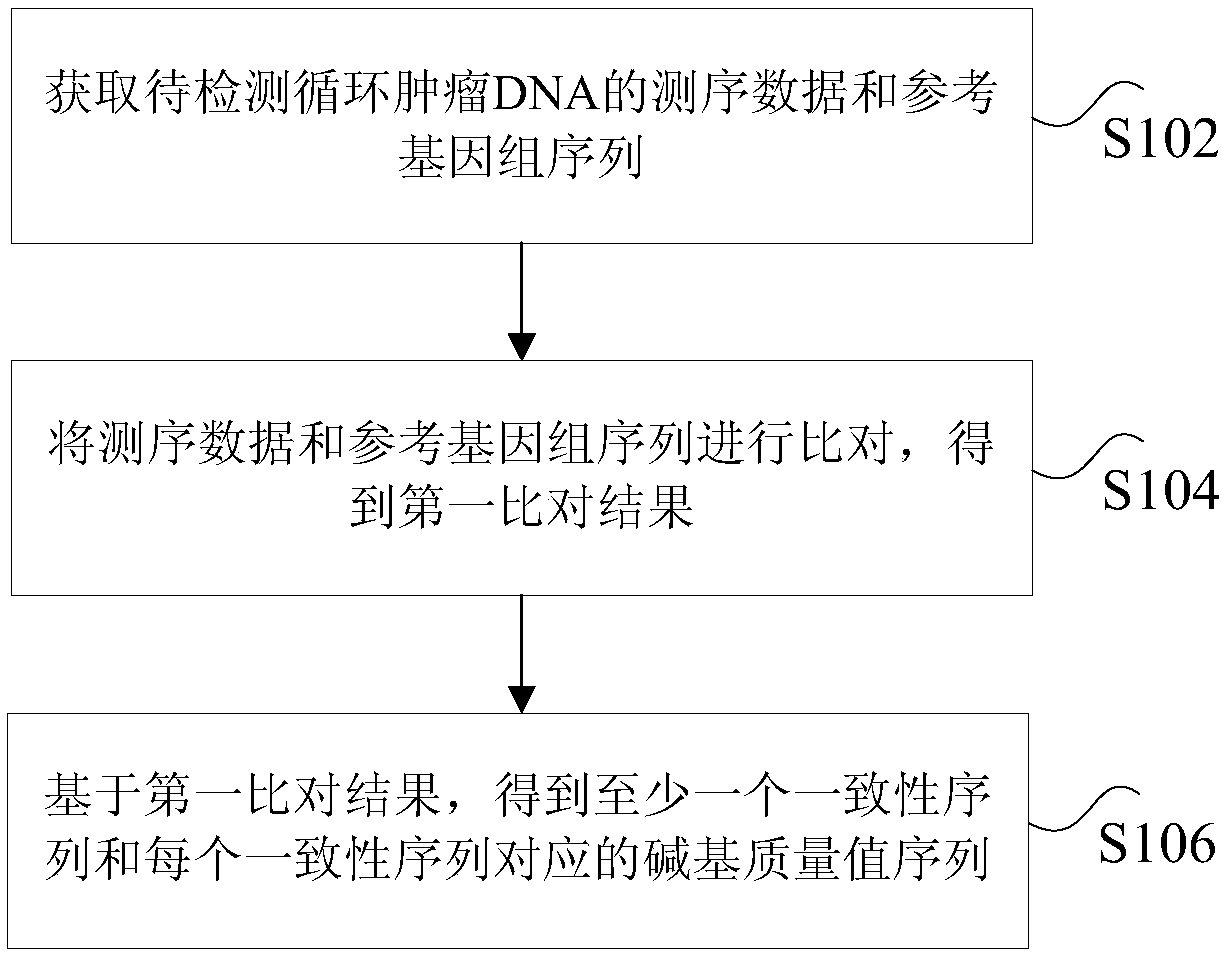
[0044] figure 1 It is a flowchart of a method for processing circulating tumor DNA repeat sequences according to an embodiment of the present invention, such as figure 1 As shown, the method includes the following steps:
[0045] Step S102, obtaining the sequencing data and the reference genome sequence of the circulating tumor DNA to be detected, wherein the sequencing data is the data obtained by high-throughput sequencing of the circulating tumor DNA to be detected, and the...
Embodiment 2
[0185] According to an embodiment of the present invention, an embodiment of a device for processing circulating tumor DNA repeat sequences is provided.
[0186] Figure 4 It is a schematic diagram of a processing device for circulating tumor DNA repeat sequences according to an embodiment of the present invention, such as Figure 4 As shown, the device includes:
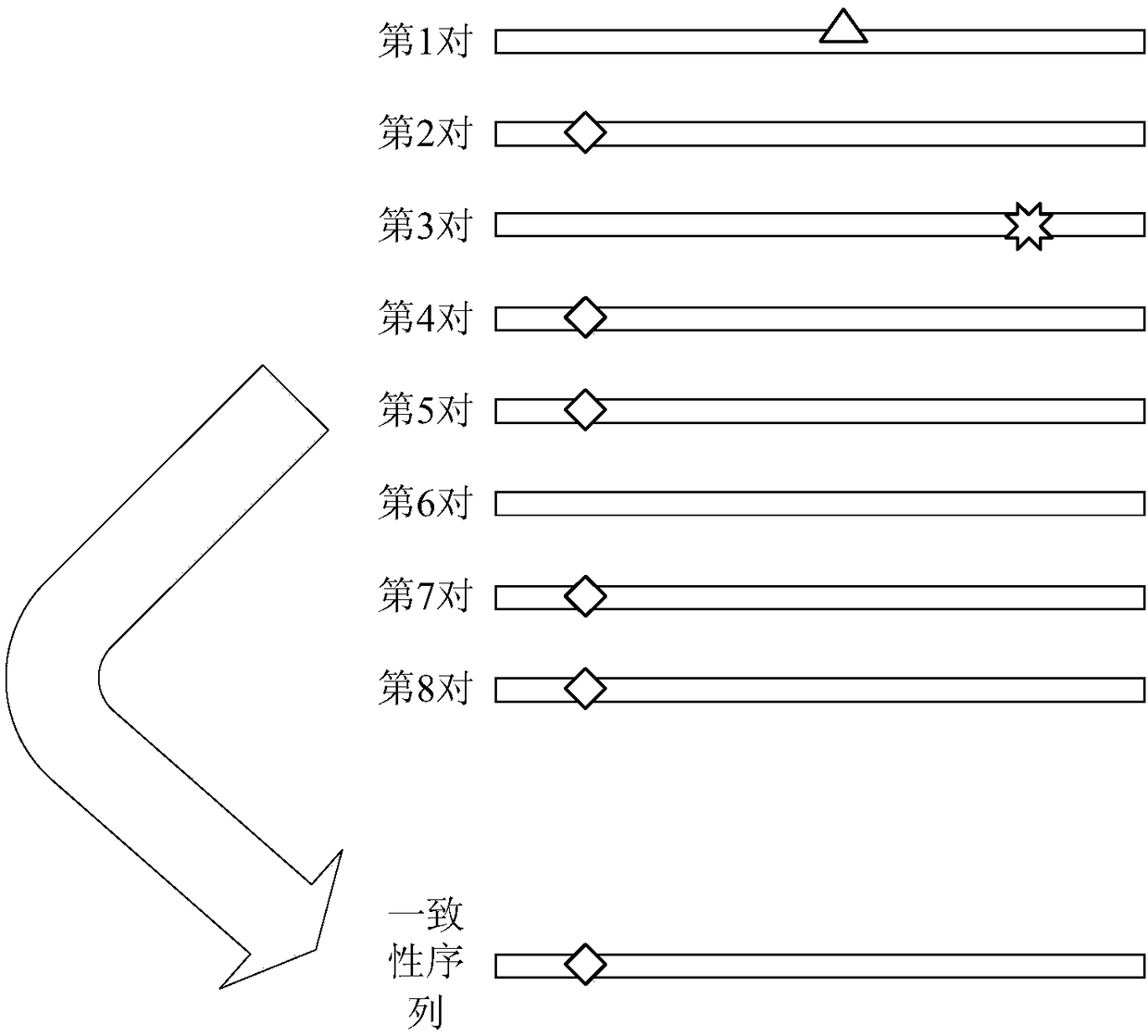
[0187] The obtaining module 42 is used to obtain the sequencing data and the reference genome sequence of the circulating tumor DNA to be detected, wherein the sequencing data is the data obtained by high-throughput sequencing of the circulating tumor DNA to be detected, and the sequencing data includes: multiple pairs of double-end sequences.
[0188] Specifically, the above-mentioned circulating tumor DNA to be detected can be the gene sequence extracted from the patient's blood, lymph fluid, interstitial fluid, cerebrospinal fluid and other body fluids. In the embodiment of the present invention, ctDNA extracted...
Embodiment 3
[0198] According to an embodiment of the present invention, an embodiment of a storage medium is provided. The storage medium includes a stored program, wherein when the program is running, the device where the storage medium is located is controlled to execute the above-mentioned method for processing circulating tumor DNA repeat sequences.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com